提高版本到yolov11
This commit is contained in:
File diff suppressed because it is too large
Load Diff
@@ -1,130 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Experimental modules."""
|
||||
|
||||
import math
|
||||
|
||||
import numpy as np
|
||||
import torch
|
||||
import torch.nn as nn
|
||||
|
||||
from utils.yolov5.utils.downloads import attempt_download
|
||||
|
||||
|
||||
class Sum(nn.Module):
|
||||
"""Weighted sum of 2 or more layers https://arxiv.org/abs/1911.09070."""
|
||||
|
||||
def __init__(self, n, weight=False):
|
||||
"""Initializes a module to sum outputs of layers with number of inputs `n` and optional weighting, supporting 2+
|
||||
inputs.
|
||||
"""
|
||||
super().__init__()
|
||||
self.weight = weight # apply weights boolean
|
||||
self.iter = range(n - 1) # iter object
|
||||
if weight:
|
||||
self.w = nn.Parameter(-torch.arange(1.0, n) / 2, requires_grad=True) # layer weights
|
||||
|
||||
def forward(self, x):
|
||||
"""Processes input through a customizable weighted sum of `n` inputs, optionally applying learned weights."""
|
||||
y = x[0] # no weight
|
||||
if self.weight:
|
||||
w = torch.sigmoid(self.w) * 2
|
||||
for i in self.iter:
|
||||
y = y + x[i + 1] * w[i]
|
||||
else:
|
||||
for i in self.iter:
|
||||
y = y + x[i + 1]
|
||||
return y
|
||||
|
||||
|
||||
class MixConv2d(nn.Module):
|
||||
"""Mixed Depth-wise Conv https://arxiv.org/abs/1907.09595."""
|
||||
|
||||
def __init__(self, c1, c2, k=(1, 3), s=1, equal_ch=True):
|
||||
"""Initializes MixConv2d with mixed depth-wise convolutional layers, taking input and output channels (c1, c2),
|
||||
kernel sizes (k), stride (s), and channel distribution strategy (equal_ch).
|
||||
"""
|
||||
super().__init__()
|
||||
n = len(k) # number of convolutions
|
||||
if equal_ch: # equal c_ per group
|
||||
i = torch.linspace(0, n - 1e-6, c2).floor() # c2 indices
|
||||
c_ = [(i == g).sum() for g in range(n)] # intermediate channels
|
||||
else: # equal weight.numel() per group
|
||||
b = [c2] + [0] * n
|
||||
a = np.eye(n + 1, n, k=-1)
|
||||
a -= np.roll(a, 1, axis=1)
|
||||
a *= np.array(k) ** 2
|
||||
a[0] = 1
|
||||
c_ = np.linalg.lstsq(a, b, rcond=None)[0].round() # solve for equal weight indices, ax = b
|
||||
|
||||
self.m = nn.ModuleList(
|
||||
[nn.Conv2d(c1, int(c_), k, s, k // 2, groups=math.gcd(c1, int(c_)), bias=False) for k, c_ in zip(k, c_)]
|
||||
)
|
||||
self.bn = nn.BatchNorm2d(c2)
|
||||
self.act = nn.SiLU()
|
||||
|
||||
def forward(self, x):
|
||||
"""Performs forward pass by applying SiLU activation on batch-normalized concatenated convolutional layer
|
||||
outputs.
|
||||
"""
|
||||
return self.act(self.bn(torch.cat([m(x) for m in self.m], 1)))
|
||||
|
||||
|
||||
class Ensemble(nn.ModuleList):
|
||||
"""Ensemble of models."""
|
||||
|
||||
def __init__(self):
|
||||
"""Initializes an ensemble of models to be used for aggregated predictions."""
|
||||
super().__init__()
|
||||
|
||||
def forward(self, x, augment=False, profile=False, visualize=False):
|
||||
"""Performs forward pass aggregating outputs from an ensemble of models.."""
|
||||
y = [module(x, augment, profile, visualize)[0] for module in self]
|
||||
# y = torch.stack(y).max(0)[0] # max ensemble
|
||||
# y = torch.stack(y).mean(0) # mean ensemble
|
||||
y = torch.cat(y, 1) # nms ensemble
|
||||
return y, None # inference, train output
|
||||
|
||||
|
||||
def attempt_load(weights, device=None, inplace=True, fuse=True):
|
||||
"""
|
||||
Loads and fuses an ensemble or single YOLOv5 model from weights, handling device placement and model adjustments.
|
||||
|
||||
Example inputs: weights=[a,b,c] or a single model weights=[a] or weights=a.
|
||||
"""
|
||||
from utils.yolov5.models.yolo import Detect, Model
|
||||
|
||||
model = Ensemble()
|
||||
for w in weights if isinstance(weights, list) else [weights]:
|
||||
ckpt = torch.load(attempt_download(w), map_location="cpu") # load
|
||||
ckpt = (ckpt.get("ema") or ckpt["model"]).to(device).float() # FP32 model
|
||||
|
||||
# Model compatibility updates
|
||||
if not hasattr(ckpt, "stride"):
|
||||
ckpt.stride = torch.tensor([32.0])
|
||||
if hasattr(ckpt, "names") and isinstance(ckpt.names, (list, tuple)):
|
||||
ckpt.names = dict(enumerate(ckpt.names)) # convert to dict
|
||||
|
||||
model.append(ckpt.fuse().eval() if fuse and hasattr(ckpt, "fuse") else ckpt.eval()) # model in eval mode
|
||||
|
||||
# Module updates
|
||||
for m in model.modules():
|
||||
t = type(m)
|
||||
if t in (nn.Hardswish, nn.LeakyReLU, nn.ReLU, nn.ReLU6, nn.SiLU, Detect, Model):
|
||||
m.inplace = inplace
|
||||
if t is Detect and not isinstance(m.anchor_grid, list):
|
||||
delattr(m, "anchor_grid")
|
||||
setattr(m, "anchor_grid", [torch.zeros(1)] * m.nl)
|
||||
elif t is nn.Upsample and not hasattr(m, "recompute_scale_factor"):
|
||||
m.recompute_scale_factor = None # torch 1.11.0 compatibility
|
||||
|
||||
# Return model
|
||||
if len(model) == 1:
|
||||
return model[-1]
|
||||
|
||||
# Return detection ensemble
|
||||
print(f"Ensemble created with {weights}\n")
|
||||
for k in "names", "nc", "yaml":
|
||||
setattr(model, k, getattr(model[0], k))
|
||||
model.stride = model[torch.argmax(torch.tensor([m.stride.max() for m in model])).int()].stride # max stride
|
||||
assert all(model[0].nc == m.nc for m in model), f"Models have different class counts: {[m.nc for m in model]}"
|
||||
return model
|
||||
@@ -1,57 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Default anchors for COCO data
|
||||
|
||||
# P5 -------------------------------------------------------------------------------------------------------------------
|
||||
# P5-640:
|
||||
anchors_p5_640:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# P6 -------------------------------------------------------------------------------------------------------------------
|
||||
# P6-640: thr=0.25: 0.9964 BPR, 5.54 anchors past thr, n=12, img_size=640, metric_all=0.281/0.716-mean/best, past_thr=0.469-mean: 9,11, 21,19, 17,41, 43,32, 39,70, 86,64, 65,131, 134,130, 120,265, 282,180, 247,354, 512,387
|
||||
anchors_p6_640:
|
||||
- [9, 11, 21, 19, 17, 41] # P3/8
|
||||
- [43, 32, 39, 70, 86, 64] # P4/16
|
||||
- [65, 131, 134, 130, 120, 265] # P5/32
|
||||
- [282, 180, 247, 354, 512, 387] # P6/64
|
||||
|
||||
# P6-1280: thr=0.25: 0.9950 BPR, 5.55 anchors past thr, n=12, img_size=1280, metric_all=0.281/0.714-mean/best, past_thr=0.468-mean: 19,27, 44,40, 38,94, 96,68, 86,152, 180,137, 140,301, 303,264, 238,542, 436,615, 739,380, 925,792
|
||||
anchors_p6_1280:
|
||||
- [19, 27, 44, 40, 38, 94] # P3/8
|
||||
- [96, 68, 86, 152, 180, 137] # P4/16
|
||||
- [140, 301, 303, 264, 238, 542] # P5/32
|
||||
- [436, 615, 739, 380, 925, 792] # P6/64
|
||||
|
||||
# P6-1920: thr=0.25: 0.9950 BPR, 5.55 anchors past thr, n=12, img_size=1920, metric_all=0.281/0.714-mean/best, past_thr=0.468-mean: 28,41, 67,59, 57,141, 144,103, 129,227, 270,205, 209,452, 455,396, 358,812, 653,922, 1109,570, 1387,1187
|
||||
anchors_p6_1920:
|
||||
- [28, 41, 67, 59, 57, 141] # P3/8
|
||||
- [144, 103, 129, 227, 270, 205] # P4/16
|
||||
- [209, 452, 455, 396, 358, 812] # P5/32
|
||||
- [653, 922, 1109, 570, 1387, 1187] # P6/64
|
||||
|
||||
# P7 -------------------------------------------------------------------------------------------------------------------
|
||||
# P7-640: thr=0.25: 0.9962 BPR, 6.76 anchors past thr, n=15, img_size=640, metric_all=0.275/0.733-mean/best, past_thr=0.466-mean: 11,11, 13,30, 29,20, 30,46, 61,38, 39,92, 78,80, 146,66, 79,163, 149,150, 321,143, 157,303, 257,402, 359,290, 524,372
|
||||
anchors_p7_640:
|
||||
- [11, 11, 13, 30, 29, 20] # P3/8
|
||||
- [30, 46, 61, 38, 39, 92] # P4/16
|
||||
- [78, 80, 146, 66, 79, 163] # P5/32
|
||||
- [149, 150, 321, 143, 157, 303] # P6/64
|
||||
- [257, 402, 359, 290, 524, 372] # P7/128
|
||||
|
||||
# P7-1280: thr=0.25: 0.9968 BPR, 6.71 anchors past thr, n=15, img_size=1280, metric_all=0.273/0.732-mean/best, past_thr=0.463-mean: 19,22, 54,36, 32,77, 70,83, 138,71, 75,173, 165,159, 148,334, 375,151, 334,317, 251,626, 499,474, 750,326, 534,814, 1079,818
|
||||
anchors_p7_1280:
|
||||
- [19, 22, 54, 36, 32, 77] # P3/8
|
||||
- [70, 83, 138, 71, 75, 173] # P4/16
|
||||
- [165, 159, 148, 334, 375, 151] # P5/32
|
||||
- [334, 317, 251, 626, 499, 474] # P6/64
|
||||
- [750, 326, 534, 814, 1079, 818] # P7/128
|
||||
|
||||
# P7-1920: thr=0.25: 0.9968 BPR, 6.71 anchors past thr, n=15, img_size=1920, metric_all=0.273/0.732-mean/best, past_thr=0.463-mean: 29,34, 81,55, 47,115, 105,124, 207,107, 113,259, 247,238, 222,500, 563,227, 501,476, 376,939, 749,711, 1126,489, 801,1222, 1618,1227
|
||||
anchors_p7_1920:
|
||||
- [29, 34, 81, 55, 47, 115] # P3/8
|
||||
- [105, 124, 207, 107, 113, 259] # P4/16
|
||||
- [247, 238, 222, 500, 563, 227] # P5/32
|
||||
- [501, 476, 376, 939, 749, 711] # P6/64
|
||||
- [1126, 489, 801, 1222, 1618, 1227] # P7/128
|
||||
@@ -1,52 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 1.0 # model depth multiple
|
||||
width_multiple: 1.0 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# darknet53 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [32, 3, 1]], # 0
|
||||
[-1, 1, Conv, [64, 3, 2]], # 1-P1/2
|
||||
[-1, 1, Bottleneck, [64]],
|
||||
[-1, 1, Conv, [128, 3, 2]], # 3-P2/4
|
||||
[-1, 2, Bottleneck, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 5-P3/8
|
||||
[-1, 8, Bottleneck, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 7-P4/16
|
||||
[-1, 8, Bottleneck, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 9-P5/32
|
||||
[-1, 4, Bottleneck, [1024]], # 10
|
||||
]
|
||||
|
||||
# YOLOv3-SPP head
|
||||
head: [
|
||||
[-1, 1, Bottleneck, [1024, False]],
|
||||
[-1, 1, SPP, [512, [5, 9, 13]]],
|
||||
[-1, 1, Conv, [1024, 3, 1]],
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, Conv, [1024, 3, 1]], # 15 (P5/32-large)
|
||||
|
||||
[-2, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 8], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 1, Bottleneck, [512, False]],
|
||||
[-1, 1, Bottleneck, [512, False]],
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, Conv, [512, 3, 1]], # 22 (P4/16-medium)
|
||||
|
||||
[-2, 1, Conv, [128, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 1, Bottleneck, [256, False]],
|
||||
[-1, 2, Bottleneck, [256, False]], # 27 (P3/8-small)
|
||||
|
||||
[[27, 22, 15], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,42 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 1.0 # model depth multiple
|
||||
width_multiple: 1.0 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 14, 23, 27, 37, 58] # P4/16
|
||||
- [81, 82, 135, 169, 344, 319] # P5/32
|
||||
|
||||
# YOLOv3-tiny backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [16, 3, 1]], # 0
|
||||
[-1, 1, nn.MaxPool2d, [2, 2, 0]], # 1-P1/2
|
||||
[-1, 1, Conv, [32, 3, 1]],
|
||||
[-1, 1, nn.MaxPool2d, [2, 2, 0]], # 3-P2/4
|
||||
[-1, 1, Conv, [64, 3, 1]],
|
||||
[-1, 1, nn.MaxPool2d, [2, 2, 0]], # 5-P3/8
|
||||
[-1, 1, Conv, [128, 3, 1]],
|
||||
[-1, 1, nn.MaxPool2d, [2, 2, 0]], # 7-P4/16
|
||||
[-1, 1, Conv, [256, 3, 1]],
|
||||
[-1, 1, nn.MaxPool2d, [2, 2, 0]], # 9-P5/32
|
||||
[-1, 1, Conv, [512, 3, 1]],
|
||||
[-1, 1, nn.ZeroPad2d, [[0, 1, 0, 1]]], # 11
|
||||
[-1, 1, nn.MaxPool2d, [2, 1, 0]], # 12
|
||||
]
|
||||
|
||||
# YOLOv3-tiny head
|
||||
head: [
|
||||
[-1, 1, Conv, [1024, 3, 1]],
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, Conv, [512, 3, 1]], # 15 (P5/32-large)
|
||||
|
||||
[-2, 1, Conv, [128, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 8], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 1, Conv, [256, 3, 1]], # 19 (P4/16-medium)
|
||||
|
||||
[[19, 15], 1, Detect, [nc, anchors]], # Detect(P4, P5)
|
||||
]
|
||||
@@ -1,52 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 1.0 # model depth multiple
|
||||
width_multiple: 1.0 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# darknet53 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [32, 3, 1]], # 0
|
||||
[-1, 1, Conv, [64, 3, 2]], # 1-P1/2
|
||||
[-1, 1, Bottleneck, [64]],
|
||||
[-1, 1, Conv, [128, 3, 2]], # 3-P2/4
|
||||
[-1, 2, Bottleneck, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 5-P3/8
|
||||
[-1, 8, Bottleneck, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 7-P4/16
|
||||
[-1, 8, Bottleneck, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 9-P5/32
|
||||
[-1, 4, Bottleneck, [1024]], # 10
|
||||
]
|
||||
|
||||
# YOLOv3 head
|
||||
head: [
|
||||
[-1, 1, Bottleneck, [1024, False]],
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, Conv, [1024, 3, 1]],
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, Conv, [1024, 3, 1]], # 15 (P5/32-large)
|
||||
|
||||
[-2, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 8], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 1, Bottleneck, [512, False]],
|
||||
[-1, 1, Bottleneck, [512, False]],
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, Conv, [512, 3, 1]], # 22 (P4/16-medium)
|
||||
|
||||
[-2, 1, Conv, [128, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 1, Bottleneck, [256, False]],
|
||||
[-1, 2, Bottleneck, [256, False]], # 27 (P3/8-small)
|
||||
|
||||
[[27, 22, 15], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,49 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 1.0 # model depth multiple
|
||||
width_multiple: 1.0 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 BiFPN head
|
||||
head: [
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 13
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 14, 6], 1, Concat, [1]], # cat P4 <--- BiFPN change
|
||||
[-1, 3, C3, [512, False]], # 20 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [1024, False]], # 23 (P5/32-large)
|
||||
|
||||
[[17, 20, 23], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,43 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 1.0 # model depth multiple
|
||||
width_multiple: 1.0 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 FPN head
|
||||
head: [
|
||||
[-1, 3, C3, [1024, False]], # 10 (P5/32-large)
|
||||
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 3, C3, [512, False]], # 14 (P4/16-medium)
|
||||
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 3, C3, [256, False]], # 18 (P3/8-small)
|
||||
|
||||
[[18, 14, 10], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,55 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 1.0 # model depth multiple
|
||||
width_multiple: 1.0 # layer channel multiple
|
||||
anchors: 3 # AutoAnchor evolves 3 anchors per P output layer
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head with (P2, P3, P4, P5) outputs
|
||||
head: [
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 13
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [128, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 2], 1, Concat, [1]], # cat backbone P2
|
||||
[-1, 1, C3, [128, False]], # 21 (P2/4-xsmall)
|
||||
|
||||
[-1, 1, Conv, [128, 3, 2]],
|
||||
[[-1, 18], 1, Concat, [1]], # cat head P3
|
||||
[-1, 3, C3, [256, False]], # 24 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 27 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [1024, False]], # 30 (P5/32-large)
|
||||
|
||||
[[21, 24, 27, 30], 1, Detect, [nc, anchors]], # Detect(P2, P3, P4, P5)
|
||||
]
|
||||
@@ -1,42 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 0.33 # model depth multiple
|
||||
width_multiple: 0.50 # layer channel multiple
|
||||
anchors: 3 # AutoAnchor evolves 3 anchors per P output layer
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head with (P3, P4) outputs
|
||||
head: [
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 13
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 20 (P4/16-medium)
|
||||
|
||||
[[17, 20], 1, Detect, [nc, anchors]], # Detect(P3, P4)
|
||||
]
|
||||
@@ -1,57 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 1.0 # model depth multiple
|
||||
width_multiple: 1.0 # layer channel multiple
|
||||
anchors: 3 # AutoAnchor evolves 3 anchors per P output layer
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [768, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [768]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 9-P6/64
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 11
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head with (P3, P4, P5, P6) outputs
|
||||
head: [
|
||||
[-1, 1, Conv, [768, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 8], 1, Concat, [1]], # cat backbone P5
|
||||
[-1, 3, C3, [768, False]], # 15
|
||||
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 19
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 23 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 20], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 26 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 16], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [768, False]], # 29 (P5/32-large)
|
||||
|
||||
[-1, 1, Conv, [768, 3, 2]],
|
||||
[[-1, 12], 1, Concat, [1]], # cat head P6
|
||||
[-1, 3, C3, [1024, False]], # 32 (P6/64-xlarge)
|
||||
|
||||
[[23, 26, 29, 32], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5, P6)
|
||||
]
|
||||
@@ -1,68 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 1.0 # model depth multiple
|
||||
width_multiple: 1.0 # layer channel multiple
|
||||
anchors: 3 # AutoAnchor evolves 3 anchors per P output layer
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [768, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [768]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 9-P6/64
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, Conv, [1280, 3, 2]], # 11-P7/128
|
||||
[-1, 3, C3, [1280]],
|
||||
[-1, 1, SPPF, [1280, 5]], # 13
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head with (P3, P4, P5, P6, P7) outputs
|
||||
head: [
|
||||
[-1, 1, Conv, [1024, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat backbone P6
|
||||
[-1, 3, C3, [1024, False]], # 17
|
||||
|
||||
[-1, 1, Conv, [768, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 8], 1, Concat, [1]], # cat backbone P5
|
||||
[-1, 3, C3, [768, False]], # 21
|
||||
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 25
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 29 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 26], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 32 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 22], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [768, False]], # 35 (P5/32-large)
|
||||
|
||||
[-1, 1, Conv, [768, 3, 2]],
|
||||
[[-1, 18], 1, Concat, [1]], # cat head P6
|
||||
[-1, 3, C3, [1024, False]], # 38 (P6/64-xlarge)
|
||||
|
||||
[-1, 1, Conv, [1024, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P7
|
||||
[-1, 3, C3, [1280, False]], # 41 (P7/128-xxlarge)
|
||||
|
||||
[[29, 32, 35, 38, 41], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5, P6, P7)
|
||||
]
|
||||
@@ -1,49 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 1.0 # model depth multiple
|
||||
width_multiple: 1.0 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 PANet head
|
||||
head: [
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 13
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 20 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [1024, False]], # 23 (P5/32-large)
|
||||
|
||||
[[17, 20, 23], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,61 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 1.0 # model depth multiple
|
||||
width_multiple: 1.0 # layer channel multiple
|
||||
anchors:
|
||||
- [19, 27, 44, 40, 38, 94] # P3/8
|
||||
- [96, 68, 86, 152, 180, 137] # P4/16
|
||||
- [140, 301, 303, 264, 238, 542] # P5/32
|
||||
- [436, 615, 739, 380, 925, 792] # P6/64
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [768, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [768]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 9-P6/64
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 11
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [768, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 8], 1, Concat, [1]], # cat backbone P5
|
||||
[-1, 3, C3, [768, False]], # 15
|
||||
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 19
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 23 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 20], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 26 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 16], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [768, False]], # 29 (P5/32-large)
|
||||
|
||||
[-1, 1, Conv, [768, 3, 2]],
|
||||
[[-1, 12], 1, Concat, [1]], # cat head P6
|
||||
[-1, 3, C3, [1024, False]], # 32 (P6/64-xlarge)
|
||||
|
||||
[[23, 26, 29, 32], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5, P6)
|
||||
]
|
||||
@@ -1,61 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 0.67 # model depth multiple
|
||||
width_multiple: 0.75 # layer channel multiple
|
||||
anchors:
|
||||
- [19, 27, 44, 40, 38, 94] # P3/8
|
||||
- [96, 68, 86, 152, 180, 137] # P4/16
|
||||
- [140, 301, 303, 264, 238, 542] # P5/32
|
||||
- [436, 615, 739, 380, 925, 792] # P6/64
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [768, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [768]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 9-P6/64
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 11
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [768, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 8], 1, Concat, [1]], # cat backbone P5
|
||||
[-1, 3, C3, [768, False]], # 15
|
||||
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 19
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 23 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 20], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 26 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 16], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [768, False]], # 29 (P5/32-large)
|
||||
|
||||
[-1, 1, Conv, [768, 3, 2]],
|
||||
[[-1, 12], 1, Concat, [1]], # cat head P6
|
||||
[-1, 3, C3, [1024, False]], # 32 (P6/64-xlarge)
|
||||
|
||||
[[23, 26, 29, 32], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5, P6)
|
||||
]
|
||||
@@ -1,61 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 0.33 # model depth multiple
|
||||
width_multiple: 0.25 # layer channel multiple
|
||||
anchors:
|
||||
- [19, 27, 44, 40, 38, 94] # P3/8
|
||||
- [96, 68, 86, 152, 180, 137] # P4/16
|
||||
- [140, 301, 303, 264, 238, 542] # P5/32
|
||||
- [436, 615, 739, 380, 925, 792] # P6/64
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [768, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [768]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 9-P6/64
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 11
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [768, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 8], 1, Concat, [1]], # cat backbone P5
|
||||
[-1, 3, C3, [768, False]], # 15
|
||||
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 19
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 23 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 20], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 26 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 16], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [768, False]], # 29 (P5/32-large)
|
||||
|
||||
[-1, 1, Conv, [768, 3, 2]],
|
||||
[[-1, 12], 1, Concat, [1]], # cat head P6
|
||||
[-1, 3, C3, [1024, False]], # 32 (P6/64-xlarge)
|
||||
|
||||
[[23, 26, 29, 32], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5, P6)
|
||||
]
|
||||
@@ -1,50 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
activation: nn.LeakyReLU(0.1) # <----- Conv() activation used throughout entire YOLOv5 model
|
||||
depth_multiple: 0.33 # model depth multiple
|
||||
width_multiple: 0.50 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 13
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 20 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [1024, False]], # 23 (P5/32-large)
|
||||
|
||||
[[17, 20, 23], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,49 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 0.33 # model depth multiple
|
||||
width_multiple: 0.50 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, GhostConv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3Ghost, [128]],
|
||||
[-1, 1, GhostConv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3Ghost, [256]],
|
||||
[-1, 1, GhostConv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3Ghost, [512]],
|
||||
[-1, 1, GhostConv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3Ghost, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, GhostConv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3Ghost, [512, False]], # 13
|
||||
|
||||
[-1, 1, GhostConv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3Ghost, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, GhostConv, [256, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3Ghost, [512, False]], # 20 (P4/16-medium)
|
||||
|
||||
[-1, 1, GhostConv, [512, 3, 2]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3Ghost, [1024, False]], # 23 (P5/32-large)
|
||||
|
||||
[[17, 20, 23], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,49 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 0.33 # model depth multiple
|
||||
width_multiple: 0.50 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3TR, [1024]], # 9 <--- C3TR() Transformer module
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 13
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 20 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [1024, False]], # 23 (P5/32-large)
|
||||
|
||||
[[17, 20, 23], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,61 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 0.33 # model depth multiple
|
||||
width_multiple: 0.50 # layer channel multiple
|
||||
anchors:
|
||||
- [19, 27, 44, 40, 38, 94] # P3/8
|
||||
- [96, 68, 86, 152, 180, 137] # P4/16
|
||||
- [140, 301, 303, 264, 238, 542] # P5/32
|
||||
- [436, 615, 739, 380, 925, 792] # P6/64
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [768, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [768]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 9-P6/64
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 11
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [768, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 8], 1, Concat, [1]], # cat backbone P5
|
||||
[-1, 3, C3, [768, False]], # 15
|
||||
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 19
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 23 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 20], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 26 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 16], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [768, False]], # 29 (P5/32-large)
|
||||
|
||||
[-1, 1, Conv, [768, 3, 2]],
|
||||
[[-1, 12], 1, Concat, [1]], # cat head P6
|
||||
[-1, 3, C3, [1024, False]], # 32 (P6/64-xlarge)
|
||||
|
||||
[[23, 26, 29, 32], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5, P6)
|
||||
]
|
||||
@@ -1,61 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 1.33 # model depth multiple
|
||||
width_multiple: 1.25 # layer channel multiple
|
||||
anchors:
|
||||
- [19, 27, 44, 40, 38, 94] # P3/8
|
||||
- [96, 68, 86, 152, 180, 137] # P4/16
|
||||
- [140, 301, 303, 264, 238, 542] # P5/32
|
||||
- [436, 615, 739, 380, 925, 792] # P6/64
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [768, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [768]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 9-P6/64
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 11
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [768, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 8], 1, Concat, [1]], # cat backbone P5
|
||||
[-1, 3, C3, [768, False]], # 15
|
||||
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 19
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 23 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 20], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 26 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 16], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [768, False]], # 29 (P5/32-large)
|
||||
|
||||
[-1, 1, Conv, [768, 3, 2]],
|
||||
[[-1, 12], 1, Concat, [1]], # cat head P6
|
||||
[-1, 3, C3, [1024, False]], # 32 (P6/64-xlarge)
|
||||
|
||||
[[23, 26, 29, 32], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5, P6)
|
||||
]
|
||||
@@ -1,49 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 1.0 # model depth multiple
|
||||
width_multiple: 1.0 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 13
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 20 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [1024, False]], # 23 (P5/32-large)
|
||||
|
||||
[[17, 20, 23], 1, Segment, [nc, anchors, 32, 256]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,49 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 0.67 # model depth multiple
|
||||
width_multiple: 0.75 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 13
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 20 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [1024, False]], # 23 (P5/32-large)
|
||||
|
||||
[[17, 20, 23], 1, Segment, [nc, anchors, 32, 256]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,49 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 0.33 # model depth multiple
|
||||
width_multiple: 0.25 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 13
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 20 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [1024, False]], # 23 (P5/32-large)
|
||||
|
||||
[[17, 20, 23], 1, Segment, [nc, anchors, 32, 256]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,49 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 0.33 # model depth multiple
|
||||
width_multiple: 0.5 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 13
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 20 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [1024, False]], # 23 (P5/32-large)
|
||||
|
||||
[[17, 20, 23], 1, Segment, [nc, anchors, 32, 256]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,49 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 1.33 # model depth multiple
|
||||
width_multiple: 1.25 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 13
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 20 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [1024, False]], # 23 (P5/32-large)
|
||||
|
||||
[[17, 20, 23], 1, Segment, [nc, anchors, 32, 256]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,797 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""
|
||||
TensorFlow, Keras and TFLite versions of YOLOv5
|
||||
Authored by https://github.com/zldrobit in PR https://github.com/ultralytics/yolov5/pull/1127.
|
||||
|
||||
Usage:
|
||||
$ python models/tf.py --weights yolov5s.pt
|
||||
|
||||
Export:
|
||||
$ python export.py --weights yolov5s.pt --include saved_model pb tflite tfjs
|
||||
"""
|
||||
|
||||
import argparse
|
||||
import sys
|
||||
from copy import deepcopy
|
||||
from pathlib import Path
|
||||
|
||||
FILE = Path(__file__).resolve()
|
||||
ROOT = FILE.parents[1] # YOLOv5 root directory
|
||||
if str(ROOT) not in sys.path:
|
||||
sys.path.append(str(ROOT)) # add ROOT to PATH
|
||||
# ROOT = ROOT.relative_to(Path.cwd()) # relative
|
||||
|
||||
import numpy as np
|
||||
import tensorflow as tf
|
||||
import torch
|
||||
import torch.nn as nn
|
||||
from tensorflow import keras
|
||||
|
||||
from models.common import (
|
||||
C3,
|
||||
SPP,
|
||||
SPPF,
|
||||
Bottleneck,
|
||||
BottleneckCSP,
|
||||
C3x,
|
||||
Concat,
|
||||
Conv,
|
||||
CrossConv,
|
||||
DWConv,
|
||||
DWConvTranspose2d,
|
||||
Focus,
|
||||
autopad,
|
||||
)
|
||||
from models.experimental import MixConv2d, attempt_load
|
||||
from models.yolo import Detect, Segment
|
||||
from utils.activations import SiLU
|
||||
from utils.general import LOGGER, make_divisible, print_args
|
||||
|
||||
|
||||
class TFBN(keras.layers.Layer):
|
||||
"""TensorFlow BatchNormalization wrapper for initializing with optional pretrained weights."""
|
||||
|
||||
def __init__(self, w=None):
|
||||
"""Initializes a TensorFlow BatchNormalization layer with optional pretrained weights."""
|
||||
super().__init__()
|
||||
self.bn = keras.layers.BatchNormalization(
|
||||
beta_initializer=keras.initializers.Constant(w.bias.numpy()),
|
||||
gamma_initializer=keras.initializers.Constant(w.weight.numpy()),
|
||||
moving_mean_initializer=keras.initializers.Constant(w.running_mean.numpy()),
|
||||
moving_variance_initializer=keras.initializers.Constant(w.running_var.numpy()),
|
||||
epsilon=w.eps,
|
||||
)
|
||||
|
||||
def call(self, inputs):
|
||||
"""Applies batch normalization to the inputs."""
|
||||
return self.bn(inputs)
|
||||
|
||||
|
||||
class TFPad(keras.layers.Layer):
|
||||
"""Pads input tensors in spatial dimensions 1 and 2 with specified integer or tuple padding values."""
|
||||
|
||||
def __init__(self, pad):
|
||||
"""
|
||||
Initializes a padding layer for spatial dimensions 1 and 2 with specified padding, supporting both int and tuple
|
||||
inputs.
|
||||
|
||||
Inputs are
|
||||
"""
|
||||
super().__init__()
|
||||
if isinstance(pad, int):
|
||||
self.pad = tf.constant([[0, 0], [pad, pad], [pad, pad], [0, 0]])
|
||||
else: # tuple/list
|
||||
self.pad = tf.constant([[0, 0], [pad[0], pad[0]], [pad[1], pad[1]], [0, 0]])
|
||||
|
||||
def call(self, inputs):
|
||||
"""Pads input tensor with zeros using specified padding, suitable for int and tuple pad dimensions."""
|
||||
return tf.pad(inputs, self.pad, mode="constant", constant_values=0)
|
||||
|
||||
|
||||
class TFConv(keras.layers.Layer):
|
||||
"""Implements a standard convolutional layer with optional batch normalization and activation for TensorFlow."""
|
||||
|
||||
def __init__(self, c1, c2, k=1, s=1, p=None, g=1, act=True, w=None):
|
||||
"""
|
||||
Initializes a standard convolution layer with optional batch normalization and activation; supports only
|
||||
group=1.
|
||||
|
||||
Inputs are ch_in, ch_out, weights, kernel, stride, padding, groups.
|
||||
"""
|
||||
super().__init__()
|
||||
assert g == 1, "TF v2.2 Conv2D does not support 'groups' argument"
|
||||
# TensorFlow convolution padding is inconsistent with PyTorch (e.g. k=3 s=2 'SAME' padding)
|
||||
# see https://stackoverflow.com/questions/52975843/comparing-conv2d-with-padding-between-tensorflow-and-pytorch
|
||||
conv = keras.layers.Conv2D(
|
||||
filters=c2,
|
||||
kernel_size=k,
|
||||
strides=s,
|
||||
padding="SAME" if s == 1 else "VALID",
|
||||
use_bias=not hasattr(w, "bn"),
|
||||
kernel_initializer=keras.initializers.Constant(w.conv.weight.permute(2, 3, 1, 0).numpy()),
|
||||
bias_initializer="zeros" if hasattr(w, "bn") else keras.initializers.Constant(w.conv.bias.numpy()),
|
||||
)
|
||||
self.conv = conv if s == 1 else keras.Sequential([TFPad(autopad(k, p)), conv])
|
||||
self.bn = TFBN(w.bn) if hasattr(w, "bn") else tf.identity
|
||||
self.act = activations(w.act) if act else tf.identity
|
||||
|
||||
def call(self, inputs):
|
||||
"""Applies convolution, batch normalization, and activation function to input tensors."""
|
||||
return self.act(self.bn(self.conv(inputs)))
|
||||
|
||||
|
||||
class TFDWConv(keras.layers.Layer):
|
||||
"""Initializes a depthwise convolution layer with optional batch normalization and activation for TensorFlow."""
|
||||
|
||||
def __init__(self, c1, c2, k=1, s=1, p=None, act=True, w=None):
|
||||
"""
|
||||
Initializes a depthwise convolution layer with optional batch normalization and activation for TensorFlow
|
||||
models.
|
||||
|
||||
Input are ch_in, ch_out, weights, kernel, stride, padding, groups.
|
||||
"""
|
||||
super().__init__()
|
||||
assert c2 % c1 == 0, f"TFDWConv() output={c2} must be a multiple of input={c1} channels"
|
||||
conv = keras.layers.DepthwiseConv2D(
|
||||
kernel_size=k,
|
||||
depth_multiplier=c2 // c1,
|
||||
strides=s,
|
||||
padding="SAME" if s == 1 else "VALID",
|
||||
use_bias=not hasattr(w, "bn"),
|
||||
depthwise_initializer=keras.initializers.Constant(w.conv.weight.permute(2, 3, 1, 0).numpy()),
|
||||
bias_initializer="zeros" if hasattr(w, "bn") else keras.initializers.Constant(w.conv.bias.numpy()),
|
||||
)
|
||||
self.conv = conv if s == 1 else keras.Sequential([TFPad(autopad(k, p)), conv])
|
||||
self.bn = TFBN(w.bn) if hasattr(w, "bn") else tf.identity
|
||||
self.act = activations(w.act) if act else tf.identity
|
||||
|
||||
def call(self, inputs):
|
||||
"""Applies convolution, batch normalization, and activation function to input tensors."""
|
||||
return self.act(self.bn(self.conv(inputs)))
|
||||
|
||||
|
||||
class TFDWConvTranspose2d(keras.layers.Layer):
|
||||
"""Implements a depthwise ConvTranspose2D layer for TensorFlow with specific settings."""
|
||||
|
||||
def __init__(self, c1, c2, k=1, s=1, p1=0, p2=0, w=None):
|
||||
"""
|
||||
Initializes depthwise ConvTranspose2D layer with specific channel, kernel, stride, and padding settings.
|
||||
|
||||
Inputs are ch_in, ch_out, weights, kernel, stride, padding, groups.
|
||||
"""
|
||||
super().__init__()
|
||||
assert c1 == c2, f"TFDWConv() output={c2} must be equal to input={c1} channels"
|
||||
assert k == 4 and p1 == 1, "TFDWConv() only valid for k=4 and p1=1"
|
||||
weight, bias = w.weight.permute(2, 3, 1, 0).numpy(), w.bias.numpy()
|
||||
self.c1 = c1
|
||||
self.conv = [
|
||||
keras.layers.Conv2DTranspose(
|
||||
filters=1,
|
||||
kernel_size=k,
|
||||
strides=s,
|
||||
padding="VALID",
|
||||
output_padding=p2,
|
||||
use_bias=True,
|
||||
kernel_initializer=keras.initializers.Constant(weight[..., i : i + 1]),
|
||||
bias_initializer=keras.initializers.Constant(bias[i]),
|
||||
)
|
||||
for i in range(c1)
|
||||
]
|
||||
|
||||
def call(self, inputs):
|
||||
"""Processes input through parallel convolutions and concatenates results, trimming border pixels."""
|
||||
return tf.concat([m(x) for m, x in zip(self.conv, tf.split(inputs, self.c1, 3))], 3)[:, 1:-1, 1:-1]
|
||||
|
||||
|
||||
class TFFocus(keras.layers.Layer):
|
||||
"""Focuses spatial information into channel space using pixel shuffling and convolution for TensorFlow models."""
|
||||
|
||||
def __init__(self, c1, c2, k=1, s=1, p=None, g=1, act=True, w=None):
|
||||
"""
|
||||
Initializes TFFocus layer to focus width and height information into channel space with custom convolution
|
||||
parameters.
|
||||
|
||||
Inputs are ch_in, ch_out, kernel, stride, padding, groups.
|
||||
"""
|
||||
super().__init__()
|
||||
self.conv = TFConv(c1 * 4, c2, k, s, p, g, act, w.conv)
|
||||
|
||||
def call(self, inputs):
|
||||
"""
|
||||
Performs pixel shuffling and convolution on input tensor, downsampling by 2 and expanding channels by 4.
|
||||
|
||||
Example x(b,w,h,c) -> y(b,w/2,h/2,4c).
|
||||
"""
|
||||
inputs = [inputs[:, ::2, ::2, :], inputs[:, 1::2, ::2, :], inputs[:, ::2, 1::2, :], inputs[:, 1::2, 1::2, :]]
|
||||
return self.conv(tf.concat(inputs, 3))
|
||||
|
||||
|
||||
class TFBottleneck(keras.layers.Layer):
|
||||
"""Implements a TensorFlow bottleneck layer with optional shortcut connections for efficient feature extraction."""
|
||||
|
||||
def __init__(self, c1, c2, shortcut=True, g=1, e=0.5, w=None):
|
||||
"""
|
||||
Initializes a standard bottleneck layer for TensorFlow models, expanding and contracting channels with optional
|
||||
shortcut.
|
||||
|
||||
Arguments are ch_in, ch_out, shortcut, groups, expansion.
|
||||
"""
|
||||
super().__init__()
|
||||
c_ = int(c2 * e) # hidden channels
|
||||
self.cv1 = TFConv(c1, c_, 1, 1, w=w.cv1)
|
||||
self.cv2 = TFConv(c_, c2, 3, 1, g=g, w=w.cv2)
|
||||
self.add = shortcut and c1 == c2
|
||||
|
||||
def call(self, inputs):
|
||||
"""Performs forward pass; if shortcut is True & input/output channels match, adds input to the convolution
|
||||
result.
|
||||
"""
|
||||
return inputs + self.cv2(self.cv1(inputs)) if self.add else self.cv2(self.cv1(inputs))
|
||||
|
||||
|
||||
class TFCrossConv(keras.layers.Layer):
|
||||
"""Implements a cross convolutional layer with optional expansion, grouping, and shortcut for TensorFlow."""
|
||||
|
||||
def __init__(self, c1, c2, k=3, s=1, g=1, e=1.0, shortcut=False, w=None):
|
||||
"""Initializes cross convolution layer with optional expansion, grouping, and shortcut addition capabilities."""
|
||||
super().__init__()
|
||||
c_ = int(c2 * e) # hidden channels
|
||||
self.cv1 = TFConv(c1, c_, (1, k), (1, s), w=w.cv1)
|
||||
self.cv2 = TFConv(c_, c2, (k, 1), (s, 1), g=g, w=w.cv2)
|
||||
self.add = shortcut and c1 == c2
|
||||
|
||||
def call(self, inputs):
|
||||
"""Passes input through two convolutions optionally adding the input if channel dimensions match."""
|
||||
return inputs + self.cv2(self.cv1(inputs)) if self.add else self.cv2(self.cv1(inputs))
|
||||
|
||||
|
||||
class TFConv2d(keras.layers.Layer):
|
||||
"""Implements a TensorFlow 2D convolution layer, mimicking PyTorch's nn.Conv2D for specified filters and stride."""
|
||||
|
||||
def __init__(self, c1, c2, k, s=1, g=1, bias=True, w=None):
|
||||
"""Initializes a TensorFlow 2D convolution layer, mimicking PyTorch's nn.Conv2D functionality for given filter
|
||||
sizes and stride.
|
||||
"""
|
||||
super().__init__()
|
||||
assert g == 1, "TF v2.2 Conv2D does not support 'groups' argument"
|
||||
self.conv = keras.layers.Conv2D(
|
||||
filters=c2,
|
||||
kernel_size=k,
|
||||
strides=s,
|
||||
padding="VALID",
|
||||
use_bias=bias,
|
||||
kernel_initializer=keras.initializers.Constant(w.weight.permute(2, 3, 1, 0).numpy()),
|
||||
bias_initializer=keras.initializers.Constant(w.bias.numpy()) if bias else None,
|
||||
)
|
||||
|
||||
def call(self, inputs):
|
||||
"""Applies a convolution operation to the inputs and returns the result."""
|
||||
return self.conv(inputs)
|
||||
|
||||
|
||||
class TFBottleneckCSP(keras.layers.Layer):
|
||||
"""Implements a CSP bottleneck layer for TensorFlow models to enhance gradient flow and efficiency."""
|
||||
|
||||
def __init__(self, c1, c2, n=1, shortcut=True, g=1, e=0.5, w=None):
|
||||
"""
|
||||
Initializes CSP bottleneck layer with specified channel sizes, count, shortcut option, groups, and expansion
|
||||
ratio.
|
||||
|
||||
Inputs are ch_in, ch_out, number, shortcut, groups, expansion.
|
||||
"""
|
||||
super().__init__()
|
||||
c_ = int(c2 * e) # hidden channels
|
||||
self.cv1 = TFConv(c1, c_, 1, 1, w=w.cv1)
|
||||
self.cv2 = TFConv2d(c1, c_, 1, 1, bias=False, w=w.cv2)
|
||||
self.cv3 = TFConv2d(c_, c_, 1, 1, bias=False, w=w.cv3)
|
||||
self.cv4 = TFConv(2 * c_, c2, 1, 1, w=w.cv4)
|
||||
self.bn = TFBN(w.bn)
|
||||
self.act = lambda x: keras.activations.swish(x)
|
||||
self.m = keras.Sequential([TFBottleneck(c_, c_, shortcut, g, e=1.0, w=w.m[j]) for j in range(n)])
|
||||
|
||||
def call(self, inputs):
|
||||
"""Processes input through the model layers, concatenates, normalizes, activates, and reduces the output
|
||||
dimensions.
|
||||
"""
|
||||
y1 = self.cv3(self.m(self.cv1(inputs)))
|
||||
y2 = self.cv2(inputs)
|
||||
return self.cv4(self.act(self.bn(tf.concat((y1, y2), axis=3))))
|
||||
|
||||
|
||||
class TFC3(keras.layers.Layer):
|
||||
"""CSP bottleneck layer with 3 convolutions for TensorFlow, supporting optional shortcuts and group convolutions."""
|
||||
|
||||
def __init__(self, c1, c2, n=1, shortcut=True, g=1, e=0.5, w=None):
|
||||
"""
|
||||
Initializes CSP Bottleneck with 3 convolutions, supporting optional shortcuts and group convolutions.
|
||||
|
||||
Inputs are ch_in, ch_out, number, shortcut, groups, expansion.
|
||||
"""
|
||||
super().__init__()
|
||||
c_ = int(c2 * e) # hidden channels
|
||||
self.cv1 = TFConv(c1, c_, 1, 1, w=w.cv1)
|
||||
self.cv2 = TFConv(c1, c_, 1, 1, w=w.cv2)
|
||||
self.cv3 = TFConv(2 * c_, c2, 1, 1, w=w.cv3)
|
||||
self.m = keras.Sequential([TFBottleneck(c_, c_, shortcut, g, e=1.0, w=w.m[j]) for j in range(n)])
|
||||
|
||||
def call(self, inputs):
|
||||
"""
|
||||
Processes input through a sequence of transformations for object detection (YOLOv5).
|
||||
|
||||
See https://github.com/ultralytics/yolov5.
|
||||
"""
|
||||
return self.cv3(tf.concat((self.m(self.cv1(inputs)), self.cv2(inputs)), axis=3))
|
||||
|
||||
|
||||
class TFC3x(keras.layers.Layer):
|
||||
"""A TensorFlow layer for enhanced feature extraction using cross-convolutions in object detection models."""
|
||||
|
||||
def __init__(self, c1, c2, n=1, shortcut=True, g=1, e=0.5, w=None):
|
||||
"""
|
||||
Initializes layer with cross-convolutions for enhanced feature extraction in object detection models.
|
||||
|
||||
Inputs are ch_in, ch_out, number, shortcut, groups, expansion.
|
||||
"""
|
||||
super().__init__()
|
||||
c_ = int(c2 * e) # hidden channels
|
||||
self.cv1 = TFConv(c1, c_, 1, 1, w=w.cv1)
|
||||
self.cv2 = TFConv(c1, c_, 1, 1, w=w.cv2)
|
||||
self.cv3 = TFConv(2 * c_, c2, 1, 1, w=w.cv3)
|
||||
self.m = keras.Sequential(
|
||||
[TFCrossConv(c_, c_, k=3, s=1, g=g, e=1.0, shortcut=shortcut, w=w.m[j]) for j in range(n)]
|
||||
)
|
||||
|
||||
def call(self, inputs):
|
||||
"""Processes input through cascaded convolutions and merges features, returning the final tensor output."""
|
||||
return self.cv3(tf.concat((self.m(self.cv1(inputs)), self.cv2(inputs)), axis=3))
|
||||
|
||||
|
||||
class TFSPP(keras.layers.Layer):
|
||||
"""Implements spatial pyramid pooling for YOLOv3-SPP with specific channels and kernel sizes."""
|
||||
|
||||
def __init__(self, c1, c2, k=(5, 9, 13), w=None):
|
||||
"""Initializes a YOLOv3-SPP layer with specific input/output channels and kernel sizes for pooling."""
|
||||
super().__init__()
|
||||
c_ = c1 // 2 # hidden channels
|
||||
self.cv1 = TFConv(c1, c_, 1, 1, w=w.cv1)
|
||||
self.cv2 = TFConv(c_ * (len(k) + 1), c2, 1, 1, w=w.cv2)
|
||||
self.m = [keras.layers.MaxPool2D(pool_size=x, strides=1, padding="SAME") for x in k]
|
||||
|
||||
def call(self, inputs):
|
||||
"""Processes input through two TFConv layers and concatenates with max-pooled outputs at intermediate stage."""
|
||||
x = self.cv1(inputs)
|
||||
return self.cv2(tf.concat([x] + [m(x) for m in self.m], 3))
|
||||
|
||||
|
||||
class TFSPPF(keras.layers.Layer):
|
||||
"""Implements a fast spatial pyramid pooling layer for TensorFlow with optimized feature extraction."""
|
||||
|
||||
def __init__(self, c1, c2, k=5, w=None):
|
||||
"""Initializes a fast spatial pyramid pooling layer with customizable in/out channels, kernel size, and
|
||||
weights.
|
||||
"""
|
||||
super().__init__()
|
||||
c_ = c1 // 2 # hidden channels
|
||||
self.cv1 = TFConv(c1, c_, 1, 1, w=w.cv1)
|
||||
self.cv2 = TFConv(c_ * 4, c2, 1, 1, w=w.cv2)
|
||||
self.m = keras.layers.MaxPool2D(pool_size=k, strides=1, padding="SAME")
|
||||
|
||||
def call(self, inputs):
|
||||
"""Executes the model's forward pass, concatenating input features with three max-pooled versions before final
|
||||
convolution.
|
||||
"""
|
||||
x = self.cv1(inputs)
|
||||
y1 = self.m(x)
|
||||
y2 = self.m(y1)
|
||||
return self.cv2(tf.concat([x, y1, y2, self.m(y2)], 3))
|
||||
|
||||
|
||||
class TFDetect(keras.layers.Layer):
|
||||
"""Implements YOLOv5 object detection layer in TensorFlow for predicting bounding boxes and class probabilities."""
|
||||
|
||||
def __init__(self, nc=80, anchors=(), ch=(), imgsz=(640, 640), w=None):
|
||||
"""Initializes YOLOv5 detection layer for TensorFlow with configurable classes, anchors, channels, and image
|
||||
size.
|
||||
"""
|
||||
super().__init__()
|
||||
self.stride = tf.convert_to_tensor(w.stride.numpy(), dtype=tf.float32)
|
||||
self.nc = nc # number of classes
|
||||
self.no = nc + 5 # number of outputs per anchor
|
||||
self.nl = len(anchors) # number of detection layers
|
||||
self.na = len(anchors[0]) // 2 # number of anchors
|
||||
self.grid = [tf.zeros(1)] * self.nl # init grid
|
||||
self.anchors = tf.convert_to_tensor(w.anchors.numpy(), dtype=tf.float32)
|
||||
self.anchor_grid = tf.reshape(self.anchors * tf.reshape(self.stride, [self.nl, 1, 1]), [self.nl, 1, -1, 1, 2])
|
||||
self.m = [TFConv2d(x, self.no * self.na, 1, w=w.m[i]) for i, x in enumerate(ch)]
|
||||
self.training = False # set to False after building model
|
||||
self.imgsz = imgsz
|
||||
for i in range(self.nl):
|
||||
ny, nx = self.imgsz[0] // self.stride[i], self.imgsz[1] // self.stride[i]
|
||||
self.grid[i] = self._make_grid(nx, ny)
|
||||
|
||||
def call(self, inputs):
|
||||
"""Performs forward pass through the model layers to predict object bounding boxes and classifications."""
|
||||
z = [] # inference output
|
||||
x = []
|
||||
for i in range(self.nl):
|
||||
x.append(self.m[i](inputs[i]))
|
||||
# x(bs,20,20,255) to x(bs,3,20,20,85)
|
||||
ny, nx = self.imgsz[0] // self.stride[i], self.imgsz[1] // self.stride[i]
|
||||
x[i] = tf.reshape(x[i], [-1, ny * nx, self.na, self.no])
|
||||
|
||||
if not self.training: # inference
|
||||
y = x[i]
|
||||
grid = tf.transpose(self.grid[i], [0, 2, 1, 3]) - 0.5
|
||||
anchor_grid = tf.transpose(self.anchor_grid[i], [0, 2, 1, 3]) * 4
|
||||
xy = (tf.sigmoid(y[..., 0:2]) * 2 + grid) * self.stride[i] # xy
|
||||
wh = tf.sigmoid(y[..., 2:4]) ** 2 * anchor_grid
|
||||
# Normalize xywh to 0-1 to reduce calibration error
|
||||
xy /= tf.constant([[self.imgsz[1], self.imgsz[0]]], dtype=tf.float32)
|
||||
wh /= tf.constant([[self.imgsz[1], self.imgsz[0]]], dtype=tf.float32)
|
||||
y = tf.concat([xy, wh, tf.sigmoid(y[..., 4 : 5 + self.nc]), y[..., 5 + self.nc :]], -1)
|
||||
z.append(tf.reshape(y, [-1, self.na * ny * nx, self.no]))
|
||||
|
||||
return tf.transpose(x, [0, 2, 1, 3]) if self.training else (tf.concat(z, 1),)
|
||||
|
||||
@staticmethod
|
||||
def _make_grid(nx=20, ny=20):
|
||||
"""Generates a 2D grid of coordinates in (x, y) format with shape [1, 1, ny*nx, 2]."""
|
||||
# return torch.stack((xv, yv), 2).view((1, 1, ny, nx, 2)).float()
|
||||
xv, yv = tf.meshgrid(tf.range(nx), tf.range(ny))
|
||||
return tf.cast(tf.reshape(tf.stack([xv, yv], 2), [1, 1, ny * nx, 2]), dtype=tf.float32)
|
||||
|
||||
|
||||
class TFSegment(TFDetect):
|
||||
"""YOLOv5 segmentation head for TensorFlow, combining detection and segmentation."""
|
||||
|
||||
def __init__(self, nc=80, anchors=(), nm=32, npr=256, ch=(), imgsz=(640, 640), w=None):
|
||||
"""Initializes YOLOv5 Segment head with specified channel depths, anchors, and input size for segmentation
|
||||
models.
|
||||
"""
|
||||
super().__init__(nc, anchors, ch, imgsz, w)
|
||||
self.nm = nm # number of masks
|
||||
self.npr = npr # number of protos
|
||||
self.no = 5 + nc + self.nm # number of outputs per anchor
|
||||
self.m = [TFConv2d(x, self.no * self.na, 1, w=w.m[i]) for i, x in enumerate(ch)] # output conv
|
||||
self.proto = TFProto(ch[0], self.npr, self.nm, w=w.proto) # protos
|
||||
self.detect = TFDetect.call
|
||||
|
||||
def call(self, x):
|
||||
"""Applies detection and proto layers on input, returning detections and optionally protos if training."""
|
||||
p = self.proto(x[0])
|
||||
# p = TFUpsample(None, scale_factor=4, mode='nearest')(self.proto(x[0])) # (optional) full-size protos
|
||||
p = tf.transpose(p, [0, 3, 1, 2]) # from shape(1,160,160,32) to shape(1,32,160,160)
|
||||
x = self.detect(self, x)
|
||||
return (x, p) if self.training else (x[0], p)
|
||||
|
||||
|
||||
class TFProto(keras.layers.Layer):
|
||||
"""Implements convolutional and upsampling layers for feature extraction in YOLOv5 segmentation."""
|
||||
|
||||
def __init__(self, c1, c_=256, c2=32, w=None):
|
||||
"""Initializes TFProto layer with convolutional and upsampling layers for feature extraction and
|
||||
transformation.
|
||||
"""
|
||||
super().__init__()
|
||||
self.cv1 = TFConv(c1, c_, k=3, w=w.cv1)
|
||||
self.upsample = TFUpsample(None, scale_factor=2, mode="nearest")
|
||||
self.cv2 = TFConv(c_, c_, k=3, w=w.cv2)
|
||||
self.cv3 = TFConv(c_, c2, w=w.cv3)
|
||||
|
||||
def call(self, inputs):
|
||||
"""Performs forward pass through the model, applying convolutions and upscaling on input tensor."""
|
||||
return self.cv3(self.cv2(self.upsample(self.cv1(inputs))))
|
||||
|
||||
|
||||
class TFUpsample(keras.layers.Layer):
|
||||
"""Implements a TensorFlow upsampling layer with specified size, scale factor, and interpolation mode."""
|
||||
|
||||
def __init__(self, size, scale_factor, mode, w=None):
|
||||
"""
|
||||
Initializes a TensorFlow upsampling layer with specified size, scale_factor, and mode, ensuring scale_factor is
|
||||
even.
|
||||
|
||||
Warning: all arguments needed including 'w'
|
||||
"""
|
||||
super().__init__()
|
||||
assert scale_factor % 2 == 0, "scale_factor must be multiple of 2"
|
||||
self.upsample = lambda x: tf.image.resize(x, (x.shape[1] * scale_factor, x.shape[2] * scale_factor), mode)
|
||||
# self.upsample = keras.layers.UpSampling2D(size=scale_factor, interpolation=mode)
|
||||
# with default arguments: align_corners=False, half_pixel_centers=False
|
||||
# self.upsample = lambda x: tf.raw_ops.ResizeNearestNeighbor(images=x,
|
||||
# size=(x.shape[1] * 2, x.shape[2] * 2))
|
||||
|
||||
def call(self, inputs):
|
||||
"""Applies upsample operation to inputs using nearest neighbor interpolation."""
|
||||
return self.upsample(inputs)
|
||||
|
||||
|
||||
class TFConcat(keras.layers.Layer):
|
||||
"""Implements TensorFlow's version of torch.concat() for concatenating tensors along the last dimension."""
|
||||
|
||||
def __init__(self, dimension=1, w=None):
|
||||
"""Initializes a TensorFlow layer for NCHW to NHWC concatenation, requiring dimension=1."""
|
||||
super().__init__()
|
||||
assert dimension == 1, "convert only NCHW to NHWC concat"
|
||||
self.d = 3
|
||||
|
||||
def call(self, inputs):
|
||||
"""Concatenates a list of tensors along the last dimension, used for NCHW to NHWC conversion."""
|
||||
return tf.concat(inputs, self.d)
|
||||
|
||||
|
||||
def parse_model(d, ch, model, imgsz):
|
||||
"""Parses a model definition dict `d` to create YOLOv5 model layers, including dynamic channel adjustments."""
|
||||
LOGGER.info(f"\n{'':>3}{'from':>18}{'n':>3}{'params':>10} {'module':<40}{'arguments':<30}")
|
||||
anchors, nc, gd, gw, ch_mul = (
|
||||
d["anchors"],
|
||||
d["nc"],
|
||||
d["depth_multiple"],
|
||||
d["width_multiple"],
|
||||
d.get("channel_multiple"),
|
||||
)
|
||||
na = (len(anchors[0]) // 2) if isinstance(anchors, list) else anchors # number of anchors
|
||||
no = na * (nc + 5) # number of outputs = anchors * (classes + 5)
|
||||
if not ch_mul:
|
||||
ch_mul = 8
|
||||
|
||||
layers, save, c2 = [], [], ch[-1] # layers, savelist, ch out
|
||||
for i, (f, n, m, args) in enumerate(d["backbone"] + d["head"]): # from, number, module, args
|
||||
m_str = m
|
||||
m = eval(m) if isinstance(m, str) else m # eval strings
|
||||
for j, a in enumerate(args):
|
||||
try:
|
||||
args[j] = eval(a) if isinstance(a, str) else a # eval strings
|
||||
except NameError:
|
||||
pass
|
||||
|
||||
n = max(round(n * gd), 1) if n > 1 else n # depth gain
|
||||
if m in [
|
||||
nn.Conv2d,
|
||||
Conv,
|
||||
DWConv,
|
||||
DWConvTranspose2d,
|
||||
Bottleneck,
|
||||
SPP,
|
||||
SPPF,
|
||||
MixConv2d,
|
||||
Focus,
|
||||
CrossConv,
|
||||
BottleneckCSP,
|
||||
C3,
|
||||
C3x,
|
||||
]:
|
||||
c1, c2 = ch[f], args[0]
|
||||
c2 = make_divisible(c2 * gw, ch_mul) if c2 != no else c2
|
||||
|
||||
args = [c1, c2, *args[1:]]
|
||||
if m in [BottleneckCSP, C3, C3x]:
|
||||
args.insert(2, n)
|
||||
n = 1
|
||||
elif m is nn.BatchNorm2d:
|
||||
args = [ch[f]]
|
||||
elif m is Concat:
|
||||
c2 = sum(ch[-1 if x == -1 else x + 1] for x in f)
|
||||
elif m in [Detect, Segment]:
|
||||
args.append([ch[x + 1] for x in f])
|
||||
if isinstance(args[1], int): # number of anchors
|
||||
args[1] = [list(range(args[1] * 2))] * len(f)
|
||||
if m is Segment:
|
||||
args[3] = make_divisible(args[3] * gw, ch_mul)
|
||||
args.append(imgsz)
|
||||
else:
|
||||
c2 = ch[f]
|
||||
|
||||
tf_m = eval("TF" + m_str.replace("nn.", ""))
|
||||
m_ = (
|
||||
keras.Sequential([tf_m(*args, w=model.model[i][j]) for j in range(n)])
|
||||
if n > 1
|
||||
else tf_m(*args, w=model.model[i])
|
||||
) # module
|
||||
|
||||
torch_m_ = nn.Sequential(*(m(*args) for _ in range(n))) if n > 1 else m(*args) # module
|
||||
t = str(m)[8:-2].replace("__main__.", "") # module type
|
||||
np = sum(x.numel() for x in torch_m_.parameters()) # number params
|
||||
m_.i, m_.f, m_.type, m_.np = i, f, t, np # attach index, 'from' index, type, number params
|
||||
LOGGER.info(f"{i:>3}{str(f):>18}{str(n):>3}{np:>10} {t:<40}{str(args):<30}") # print
|
||||
save.extend(x % i for x in ([f] if isinstance(f, int) else f) if x != -1) # append to savelist
|
||||
layers.append(m_)
|
||||
ch.append(c2)
|
||||
return keras.Sequential(layers), sorted(save)
|
||||
|
||||
|
||||
class TFModel:
|
||||
"""Implements YOLOv5 model in TensorFlow, supporting TensorFlow, Keras, and TFLite formats for object detection."""
|
||||
|
||||
def __init__(self, cfg="yolov5s.yaml", ch=3, nc=None, model=None, imgsz=(640, 640)):
|
||||
"""Initializes TF YOLOv5 model with specified configuration, channels, classes, model instance, and input
|
||||
size.
|
||||
"""
|
||||
super().__init__()
|
||||
if isinstance(cfg, dict):
|
||||
self.yaml = cfg # model dict
|
||||
else: # is *.yaml
|
||||
import yaml # for torch hub
|
||||
|
||||
self.yaml_file = Path(cfg).name
|
||||
with open(cfg) as f:
|
||||
self.yaml = yaml.load(f, Loader=yaml.FullLoader) # model dict
|
||||
|
||||
# Define model
|
||||
if nc and nc != self.yaml["nc"]:
|
||||
LOGGER.info(f"Overriding {cfg} nc={self.yaml['nc']} with nc={nc}")
|
||||
self.yaml["nc"] = nc # override yaml value
|
||||
self.model, self.savelist = parse_model(deepcopy(self.yaml), ch=[ch], model=model, imgsz=imgsz)
|
||||
|
||||
def predict(
|
||||
self,
|
||||
inputs,
|
||||
tf_nms=False,
|
||||
agnostic_nms=False,
|
||||
topk_per_class=100,
|
||||
topk_all=100,
|
||||
iou_thres=0.45,
|
||||
conf_thres=0.25,
|
||||
):
|
||||
"""Runs inference on input data, with an option for TensorFlow NMS."""
|
||||
y = [] # outputs
|
||||
x = inputs
|
||||
for m in self.model.layers:
|
||||
if m.f != -1: # if not from previous layer
|
||||
x = y[m.f] if isinstance(m.f, int) else [x if j == -1 else y[j] for j in m.f] # from earlier layers
|
||||
|
||||
x = m(x) # run
|
||||
y.append(x if m.i in self.savelist else None) # save output
|
||||
|
||||
# Add TensorFlow NMS
|
||||
if tf_nms:
|
||||
boxes = self._xywh2xyxy(x[0][..., :4])
|
||||
probs = x[0][:, :, 4:5]
|
||||
classes = x[0][:, :, 5:]
|
||||
scores = probs * classes
|
||||
if agnostic_nms:
|
||||
nms = AgnosticNMS()((boxes, classes, scores), topk_all, iou_thres, conf_thres)
|
||||
else:
|
||||
boxes = tf.expand_dims(boxes, 2)
|
||||
nms = tf.image.combined_non_max_suppression(
|
||||
boxes, scores, topk_per_class, topk_all, iou_thres, conf_thres, clip_boxes=False
|
||||
)
|
||||
return (nms,)
|
||||
return x # output [1,6300,85] = [xywh, conf, class0, class1, ...]
|
||||
# x = x[0] # [x(1,6300,85), ...] to x(6300,85)
|
||||
# xywh = x[..., :4] # x(6300,4) boxes
|
||||
# conf = x[..., 4:5] # x(6300,1) confidences
|
||||
# cls = tf.reshape(tf.cast(tf.argmax(x[..., 5:], axis=1), tf.float32), (-1, 1)) # x(6300,1) classes
|
||||
# return tf.concat([conf, cls, xywh], 1)
|
||||
|
||||
@staticmethod
|
||||
def _xywh2xyxy(xywh):
|
||||
"""Converts bounding box format from [x, y, w, h] to [x1, y1, x2, y2], where xy1=top-left and xy2=bottom-
|
||||
right.
|
||||
"""
|
||||
x, y, w, h = tf.split(xywh, num_or_size_splits=4, axis=-1)
|
||||
return tf.concat([x - w / 2, y - h / 2, x + w / 2, y + h / 2], axis=-1)
|
||||
|
||||
|
||||
class AgnosticNMS(keras.layers.Layer):
|
||||
"""Performs agnostic non-maximum suppression (NMS) on detected objects using IoU and confidence thresholds."""
|
||||
|
||||
def call(self, input, topk_all, iou_thres, conf_thres):
|
||||
"""Performs agnostic NMS on input tensors using given thresholds and top-K selection."""
|
||||
return tf.map_fn(
|
||||
lambda x: self._nms(x, topk_all, iou_thres, conf_thres),
|
||||
input,
|
||||
fn_output_signature=(tf.float32, tf.float32, tf.float32, tf.int32),
|
||||
name="agnostic_nms",
|
||||
)
|
||||
|
||||
@staticmethod
|
||||
def _nms(x, topk_all=100, iou_thres=0.45, conf_thres=0.25):
|
||||
"""Performs agnostic non-maximum suppression (NMS) on detected objects, filtering based on IoU and confidence
|
||||
thresholds.
|
||||
"""
|
||||
boxes, classes, scores = x
|
||||
class_inds = tf.cast(tf.argmax(classes, axis=-1), tf.float32)
|
||||
scores_inp = tf.reduce_max(scores, -1)
|
||||
selected_inds = tf.image.non_max_suppression(
|
||||
boxes, scores_inp, max_output_size=topk_all, iou_threshold=iou_thres, score_threshold=conf_thres
|
||||
)
|
||||
selected_boxes = tf.gather(boxes, selected_inds)
|
||||
padded_boxes = tf.pad(
|
||||
selected_boxes,
|
||||
paddings=[[0, topk_all - tf.shape(selected_boxes)[0]], [0, 0]],
|
||||
mode="CONSTANT",
|
||||
constant_values=0.0,
|
||||
)
|
||||
selected_scores = tf.gather(scores_inp, selected_inds)
|
||||
padded_scores = tf.pad(
|
||||
selected_scores,
|
||||
paddings=[[0, topk_all - tf.shape(selected_boxes)[0]]],
|
||||
mode="CONSTANT",
|
||||
constant_values=-1.0,
|
||||
)
|
||||
selected_classes = tf.gather(class_inds, selected_inds)
|
||||
padded_classes = tf.pad(
|
||||
selected_classes,
|
||||
paddings=[[0, topk_all - tf.shape(selected_boxes)[0]]],
|
||||
mode="CONSTANT",
|
||||
constant_values=-1.0,
|
||||
)
|
||||
valid_detections = tf.shape(selected_inds)[0]
|
||||
return padded_boxes, padded_scores, padded_classes, valid_detections
|
||||
|
||||
|
||||
def activations(act=nn.SiLU):
|
||||
"""Converts PyTorch activations to TensorFlow equivalents, supporting LeakyReLU, Hardswish, and SiLU/Swish."""
|
||||
if isinstance(act, nn.LeakyReLU):
|
||||
return lambda x: keras.activations.relu(x, alpha=0.1)
|
||||
elif isinstance(act, nn.Hardswish):
|
||||
return lambda x: x * tf.nn.relu6(x + 3) * 0.166666667
|
||||
elif isinstance(act, (nn.SiLU, SiLU)):
|
||||
return lambda x: keras.activations.swish(x)
|
||||
else:
|
||||
raise Exception(f"no matching TensorFlow activation found for PyTorch activation {act}")
|
||||
|
||||
|
||||
def representative_dataset_gen(dataset, ncalib=100):
|
||||
"""Generates a representative dataset for calibration by yielding transformed numpy arrays from the input
|
||||
dataset.
|
||||
"""
|
||||
for n, (path, img, im0s, vid_cap, string) in enumerate(dataset):
|
||||
im = np.transpose(img, [1, 2, 0])
|
||||
im = np.expand_dims(im, axis=0).astype(np.float32)
|
||||
im /= 255
|
||||
yield [im]
|
||||
if n >= ncalib:
|
||||
break
|
||||
|
||||
|
||||
def run(
|
||||
weights=ROOT / "yolov5s.pt", # weights path
|
||||
imgsz=(640, 640), # inference size h,w
|
||||
batch_size=1, # batch size
|
||||
dynamic=False, # dynamic batch size
|
||||
):
|
||||
# PyTorch model
|
||||
"""Exports YOLOv5 model from PyTorch to TensorFlow and Keras formats, performing inference for validation."""
|
||||
im = torch.zeros((batch_size, 3, *imgsz)) # BCHW image
|
||||
model = attempt_load(weights, device=torch.device("cpu"), inplace=True, fuse=False)
|
||||
_ = model(im) # inference
|
||||
model.info()
|
||||
|
||||
# TensorFlow model
|
||||
im = tf.zeros((batch_size, *imgsz, 3)) # BHWC image
|
||||
tf_model = TFModel(cfg=model.yaml, model=model, nc=model.nc, imgsz=imgsz)
|
||||
_ = tf_model.predict(im) # inference
|
||||
|
||||
# Keras model
|
||||
im = keras.Input(shape=(*imgsz, 3), batch_size=None if dynamic else batch_size)
|
||||
keras_model = keras.Model(inputs=im, outputs=tf_model.predict(im))
|
||||
keras_model.summary()
|
||||
|
||||
LOGGER.info("PyTorch, TensorFlow and Keras models successfully verified.\nUse export.py for TF model export.")
|
||||
|
||||
|
||||
def parse_opt():
|
||||
"""Parses and returns command-line options for model inference, including weights path, image size, batch size, and
|
||||
dynamic batching.
|
||||
"""
|
||||
parser = argparse.ArgumentParser()
|
||||
parser.add_argument("--weights", type=str, default=ROOT / "yolov5s.pt", help="weights path")
|
||||
parser.add_argument("--imgsz", "--img", "--img-size", nargs="+", type=int, default=[640], help="inference size h,w")
|
||||
parser.add_argument("--batch-size", type=int, default=1, help="batch size")
|
||||
parser.add_argument("--dynamic", action="store_true", help="dynamic batch size")
|
||||
opt = parser.parse_args()
|
||||
opt.imgsz *= 2 if len(opt.imgsz) == 1 else 1 # expand
|
||||
print_args(vars(opt))
|
||||
return opt
|
||||
|
||||
|
||||
def main(opt):
|
||||
"""Executes the YOLOv5 model run function with parsed command line options."""
|
||||
run(**vars(opt))
|
||||
|
||||
|
||||
if __name__ == "__main__":
|
||||
opt = parse_opt()
|
||||
main(opt)
|
||||
@@ -1,495 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""
|
||||
YOLO-specific modules.
|
||||
|
||||
Usage:
|
||||
$ python models/yolo.py --cfg yolov5s.yaml
|
||||
"""
|
||||
|
||||
import argparse
|
||||
import contextlib
|
||||
import math
|
||||
import os
|
||||
import platform
|
||||
import sys
|
||||
from copy import deepcopy
|
||||
from pathlib import Path
|
||||
|
||||
import torch
|
||||
import torch.nn as nn
|
||||
|
||||
FILE = Path(__file__).resolve()
|
||||
ROOT = FILE.parents[1] # YOLOv5 root directory
|
||||
if str(ROOT) not in sys.path:
|
||||
sys.path.append(str(ROOT)) # add ROOT to PATH
|
||||
if platform.system() != "Windows":
|
||||
ROOT = Path(os.path.relpath(ROOT, Path.cwd())) # relative
|
||||
|
||||
from utils.yolov5.models.common import (
|
||||
C3,
|
||||
C3SPP,
|
||||
C3TR,
|
||||
SPP,
|
||||
SPPF,
|
||||
Bottleneck,
|
||||
BottleneckCSP,
|
||||
C3Ghost,
|
||||
C3x,
|
||||
Classify,
|
||||
Concat,
|
||||
Contract,
|
||||
Conv,
|
||||
CrossConv,
|
||||
DetectMultiBackend,
|
||||
DWConv,
|
||||
DWConvTranspose2d,
|
||||
Expand,
|
||||
Focus,
|
||||
GhostBottleneck,
|
||||
GhostConv,
|
||||
Proto,
|
||||
)
|
||||
from utils.yolov5.models.experimental import MixConv2d
|
||||
from utils.yolov5.utils.autoanchor import check_anchor_order
|
||||
from utils.yolov5.utils.general import LOGGER, check_version, check_yaml, colorstr, make_divisible, print_args
|
||||
from utils.yolov5.utils.plots import feature_visualization
|
||||
from utils.yolov5.utils.torch_utils import (
|
||||
fuse_conv_and_bn,
|
||||
initialize_weights,
|
||||
model_info,
|
||||
profile,
|
||||
scale_img,
|
||||
select_device,
|
||||
time_sync,
|
||||
)
|
||||
|
||||
try:
|
||||
import thop # for FLOPs computation
|
||||
except ImportError:
|
||||
thop = None
|
||||
|
||||
|
||||
class Detect(nn.Module):
|
||||
"""YOLOv5 Detect head for processing input tensors and generating detection outputs in object detection models."""
|
||||
|
||||
stride = None # strides computed during build
|
||||
dynamic = False # force grid reconstruction
|
||||
export = False # export mode
|
||||
|
||||
def __init__(self, nc=80, anchors=(), ch=(), inplace=True):
|
||||
"""Initializes YOLOv5 detection layer with specified classes, anchors, channels, and inplace operations."""
|
||||
super().__init__()
|
||||
self.nc = nc # number of classes
|
||||
self.no = nc + 5 # number of outputs per anchor
|
||||
self.nl = len(anchors) # number of detection layers
|
||||
self.na = len(anchors[0]) // 2 # number of anchors
|
||||
self.grid = [torch.empty(0) for _ in range(self.nl)] # init grid
|
||||
self.anchor_grid = [torch.empty(0) for _ in range(self.nl)] # init anchor grid
|
||||
self.register_buffer("anchors", torch.tensor(anchors).float().view(self.nl, -1, 2)) # shape(nl,na,2)
|
||||
self.m = nn.ModuleList(nn.Conv2d(x, self.no * self.na, 1) for x in ch) # output conv
|
||||
self.inplace = inplace # use inplace ops (e.g. slice assignment)
|
||||
|
||||
def forward(self, x):
|
||||
"""Processes input through YOLOv5 layers, altering shape for detection: `x(bs, 3, ny, nx, 85)`."""
|
||||
z = [] # inference output
|
||||
for i in range(self.nl):
|
||||
x[i] = self.m[i](x[i]) # conv
|
||||
bs, _, ny, nx = x[i].shape # x(bs,255,20,20) to x(bs,3,20,20,85)
|
||||
x[i] = x[i].view(bs, self.na, self.no, ny, nx).permute(0, 1, 3, 4, 2).contiguous()
|
||||
|
||||
if not self.training: # inference
|
||||
if self.dynamic or self.grid[i].shape[2:4] != x[i].shape[2:4]:
|
||||
self.grid[i], self.anchor_grid[i] = self._make_grid(nx, ny, i)
|
||||
|
||||
if isinstance(self, Segment): # (boxes + masks)
|
||||
xy, wh, conf, mask = x[i].split((2, 2, self.nc + 1, self.no - self.nc - 5), 4)
|
||||
xy = (xy.sigmoid() * 2 + self.grid[i]) * self.stride[i] # xy
|
||||
wh = (wh.sigmoid() * 2) ** 2 * self.anchor_grid[i] # wh
|
||||
y = torch.cat((xy, wh, conf.sigmoid(), mask), 4)
|
||||
else: # Detect (boxes only)
|
||||
xy, wh, conf = x[i].sigmoid().split((2, 2, self.nc + 1), 4)
|
||||
xy = (xy * 2 + self.grid[i]) * self.stride[i] # xy
|
||||
wh = (wh * 2) ** 2 * self.anchor_grid[i] # wh
|
||||
y = torch.cat((xy, wh, conf), 4)
|
||||
z.append(y.view(bs, self.na * nx * ny, self.no))
|
||||
|
||||
return x if self.training else (torch.cat(z, 1),) if self.export else (torch.cat(z, 1), x)
|
||||
|
||||
def _make_grid(self, nx=20, ny=20, i=0, torch_1_10=check_version(torch.__version__, "1.10.0")):
|
||||
"""Generates a mesh grid for anchor boxes with optional compatibility for torch versions < 1.10."""
|
||||
d = self.anchors[i].device
|
||||
t = self.anchors[i].dtype
|
||||
shape = 1, self.na, ny, nx, 2 # grid shape
|
||||
y, x = torch.arange(ny, device=d, dtype=t), torch.arange(nx, device=d, dtype=t)
|
||||
yv, xv = torch.meshgrid(y, x, indexing="ij") if torch_1_10 else torch.meshgrid(y, x) # torch>=0.7 compatibility
|
||||
grid = torch.stack((xv, yv), 2).expand(shape) - 0.5 # add grid offset, i.e. y = 2.0 * x - 0.5
|
||||
anchor_grid = (self.anchors[i] * self.stride[i]).view((1, self.na, 1, 1, 2)).expand(shape)
|
||||
return grid, anchor_grid
|
||||
|
||||
|
||||
class Segment(Detect):
|
||||
"""YOLOv5 Segment head for segmentation models, extending Detect with mask and prototype layers."""
|
||||
|
||||
def __init__(self, nc=80, anchors=(), nm=32, npr=256, ch=(), inplace=True):
|
||||
"""Initializes YOLOv5 Segment head with options for mask count, protos, and channel adjustments."""
|
||||
super().__init__(nc, anchors, ch, inplace)
|
||||
self.nm = nm # number of masks
|
||||
self.npr = npr # number of protos
|
||||
self.no = 5 + nc + self.nm # number of outputs per anchor
|
||||
self.m = nn.ModuleList(nn.Conv2d(x, self.no * self.na, 1) for x in ch) # output conv
|
||||
self.proto = Proto(ch[0], self.npr, self.nm) # protos
|
||||
self.detect = Detect.forward
|
||||
|
||||
def forward(self, x):
|
||||
"""Processes input through the network, returning detections and prototypes; adjusts output based on
|
||||
training/export mode.
|
||||
"""
|
||||
p = self.proto(x[0])
|
||||
x = self.detect(self, x)
|
||||
return (x, p) if self.training else (x[0], p) if self.export else (x[0], p, x[1])
|
||||
|
||||
|
||||
class BaseModel(nn.Module):
|
||||
"""YOLOv5 base model."""
|
||||
|
||||
def forward(self, x, profile=False, visualize=False):
|
||||
"""Executes a single-scale inference or training pass on the YOLOv5 base model, with options for profiling and
|
||||
visualization.
|
||||
"""
|
||||
return self._forward_once(x, profile, visualize) # single-scale inference, train
|
||||
|
||||
def _forward_once(self, x, profile=False, visualize=False):
|
||||
"""Performs a forward pass on the YOLOv5 model, enabling profiling and feature visualization options."""
|
||||
y, dt = [], [] # outputs
|
||||
for m in self.model:
|
||||
if m.f != -1: # if not from previous layer
|
||||
x = y[m.f] if isinstance(m.f, int) else [x if j == -1 else y[j] for j in m.f] # from earlier layers
|
||||
if profile:
|
||||
self._profile_one_layer(m, x, dt)
|
||||
x = m(x) # run
|
||||
y.append(x if m.i in self.save else None) # save output
|
||||
if visualize:
|
||||
feature_visualization(x, m.type, m.i, save_dir=visualize)
|
||||
return x
|
||||
|
||||
def _profile_one_layer(self, m, x, dt):
|
||||
"""Profiles a single layer's performance by computing GFLOPs, execution time, and parameters."""
|
||||
c = m == self.model[-1] # is final layer, copy input as inplace fix
|
||||
o = thop.profile(m, inputs=(x.copy() if c else x,), verbose=False)[0] / 1e9 * 2 if thop else 0 # FLOPs
|
||||
t = time_sync()
|
||||
for _ in range(10):
|
||||
m(x.copy() if c else x)
|
||||
dt.append((time_sync() - t) * 100)
|
||||
if m == self.model[0]:
|
||||
LOGGER.info(f"{'time (ms)':>10s} {'GFLOPs':>10s} {'params':>10s} module")
|
||||
LOGGER.info(f"{dt[-1]:10.2f} {o:10.2f} {m.np:10.0f} {m.type}")
|
||||
if c:
|
||||
LOGGER.info(f"{sum(dt):10.2f} {'-':>10s} {'-':>10s} Total")
|
||||
|
||||
def fuse(self):
|
||||
"""Fuses Conv2d() and BatchNorm2d() layers in the model to improve inference speed."""
|
||||
LOGGER.info("Fusing layers... ")
|
||||
for m in self.model.modules():
|
||||
if isinstance(m, (Conv, DWConv)) and hasattr(m, "bn"):
|
||||
m.conv = fuse_conv_and_bn(m.conv, m.bn) # update conv
|
||||
delattr(m, "bn") # remove batchnorm
|
||||
m.forward = m.forward_fuse # update forward
|
||||
self.info()
|
||||
return self
|
||||
|
||||
def info(self, verbose=False, img_size=640):
|
||||
"""Prints model information given verbosity and image size, e.g., `info(verbose=True, img_size=640)`."""
|
||||
model_info(self, verbose, img_size)
|
||||
|
||||
def _apply(self, fn):
|
||||
"""Applies transformations like to(), cpu(), cuda(), half() to model tensors excluding parameters or registered
|
||||
buffers.
|
||||
"""
|
||||
self = super()._apply(fn)
|
||||
m = self.model[-1] # Detect()
|
||||
if isinstance(m, (Detect, Segment)):
|
||||
m.stride = fn(m.stride)
|
||||
m.grid = list(map(fn, m.grid))
|
||||
if isinstance(m.anchor_grid, list):
|
||||
m.anchor_grid = list(map(fn, m.anchor_grid))
|
||||
return self
|
||||
|
||||
|
||||
class DetectionModel(BaseModel):
|
||||
"""YOLOv5 detection model class for object detection tasks, supporting custom configurations and anchors."""
|
||||
|
||||
def __init__(self, cfg="yolov5s.yaml", ch=3, nc=None, anchors=None):
|
||||
"""Initializes YOLOv5 model with configuration file, input channels, number of classes, and custom anchors."""
|
||||
super().__init__()
|
||||
if isinstance(cfg, dict):
|
||||
self.yaml = cfg # model dict
|
||||
else: # is *.yaml
|
||||
import yaml # for torch hub
|
||||
|
||||
self.yaml_file = Path(cfg).name
|
||||
with open(cfg, encoding="ascii", errors="ignore") as f:
|
||||
self.yaml = yaml.safe_load(f) # model dict
|
||||
|
||||
# Define model
|
||||
ch = self.yaml["ch"] = self.yaml.get("ch", ch) # input channels
|
||||
if nc and nc != self.yaml["nc"]:
|
||||
LOGGER.info(f"Overriding model.yaml nc={self.yaml['nc']} with nc={nc}")
|
||||
self.yaml["nc"] = nc # override yaml value
|
||||
if anchors:
|
||||
LOGGER.info(f"Overriding model.yaml anchors with anchors={anchors}")
|
||||
self.yaml["anchors"] = round(anchors) # override yaml value
|
||||
self.model, self.save = parse_model(deepcopy(self.yaml), ch=[ch]) # model, savelist
|
||||
self.names = [str(i) for i in range(self.yaml["nc"])] # default names
|
||||
self.inplace = self.yaml.get("inplace", True)
|
||||
|
||||
# Build strides, anchors
|
||||
m = self.model[-1] # Detect()
|
||||
if isinstance(m, (Detect, Segment)):
|
||||
|
||||
def _forward(x):
|
||||
"""Passes the input 'x' through the model and returns the processed output."""
|
||||
return self.forward(x)[0] if isinstance(m, Segment) else self.forward(x)
|
||||
|
||||
s = 256 # 2x min stride
|
||||
m.inplace = self.inplace
|
||||
m.stride = torch.tensor([s / x.shape[-2] for x in _forward(torch.zeros(1, ch, s, s))]) # forward
|
||||
check_anchor_order(m)
|
||||
m.anchors /= m.stride.view(-1, 1, 1)
|
||||
self.stride = m.stride
|
||||
self._initialize_biases() # only run once
|
||||
|
||||
# Init weights, biases
|
||||
initialize_weights(self)
|
||||
self.info()
|
||||
LOGGER.info("")
|
||||
|
||||
def forward(self, x, augment=False, profile=False, visualize=False):
|
||||
"""Performs single-scale or augmented inference and may include profiling or visualization."""
|
||||
if augment:
|
||||
return self._forward_augment(x) # augmented inference, None
|
||||
return self._forward_once(x, profile, visualize) # single-scale inference, train
|
||||
|
||||
def _forward_augment(self, x):
|
||||
"""Performs augmented inference across different scales and flips, returning combined detections."""
|
||||
img_size = x.shape[-2:] # height, width
|
||||
s = [1, 0.83, 0.67] # scales
|
||||
f = [None, 3, None] # flips (2-ud, 3-lr)
|
||||
y = [] # outputs
|
||||
for si, fi in zip(s, f):
|
||||
xi = scale_img(x.flip(fi) if fi else x, si, gs=int(self.stride.max()))
|
||||
yi = self._forward_once(xi)[0] # forward
|
||||
# cv2.imwrite(f'img_{si}.jpg', 255 * xi[0].cpu().numpy().transpose((1, 2, 0))[:, :, ::-1]) # save
|
||||
yi = self._descale_pred(yi, fi, si, img_size)
|
||||
y.append(yi)
|
||||
y = self._clip_augmented(y) # clip augmented tails
|
||||
return torch.cat(y, 1), None # augmented inference, train
|
||||
|
||||
def _descale_pred(self, p, flips, scale, img_size):
|
||||
"""De-scales predictions from augmented inference, adjusting for flips and image size."""
|
||||
if self.inplace:
|
||||
p[..., :4] /= scale # de-scale
|
||||
if flips == 2:
|
||||
p[..., 1] = img_size[0] - p[..., 1] # de-flip ud
|
||||
elif flips == 3:
|
||||
p[..., 0] = img_size[1] - p[..., 0] # de-flip lr
|
||||
else:
|
||||
x, y, wh = p[..., 0:1] / scale, p[..., 1:2] / scale, p[..., 2:4] / scale # de-scale
|
||||
if flips == 2:
|
||||
y = img_size[0] - y # de-flip ud
|
||||
elif flips == 3:
|
||||
x = img_size[1] - x # de-flip lr
|
||||
p = torch.cat((x, y, wh, p[..., 4:]), -1)
|
||||
return p
|
||||
|
||||
def _clip_augmented(self, y):
|
||||
"""Clips augmented inference tails for YOLOv5 models, affecting first and last tensors based on grid points and
|
||||
layer counts.
|
||||
"""
|
||||
nl = self.model[-1].nl # number of detection layers (P3-P5)
|
||||
g = sum(4**x for x in range(nl)) # grid points
|
||||
e = 1 # exclude layer count
|
||||
i = (y[0].shape[1] // g) * sum(4**x for x in range(e)) # indices
|
||||
y[0] = y[0][:, :-i] # large
|
||||
i = (y[-1].shape[1] // g) * sum(4 ** (nl - 1 - x) for x in range(e)) # indices
|
||||
y[-1] = y[-1][:, i:] # small
|
||||
return y
|
||||
|
||||
def _initialize_biases(self, cf=None):
|
||||
"""
|
||||
Initializes biases for YOLOv5's Detect() module, optionally using class frequencies (cf).
|
||||
|
||||
For details see https://arxiv.org/abs/1708.02002 section 3.3.
|
||||
"""
|
||||
# cf = torch.bincount(torch.tensor(np.concatenate(dataset.labels, 0)[:, 0]).long(), minlength=nc) + 1.
|
||||
m = self.model[-1] # Detect() module
|
||||
for mi, s in zip(m.m, m.stride): # from
|
||||
b = mi.bias.view(m.na, -1) # conv.bias(255) to (3,85)
|
||||
b.data[:, 4] += math.log(8 / (640 / s) ** 2) # obj (8 objects per 640 image)
|
||||
b.data[:, 5 : 5 + m.nc] += (
|
||||
math.log(0.6 / (m.nc - 0.99999)) if cf is None else torch.log(cf / cf.sum())
|
||||
) # cls
|
||||
mi.bias = torch.nn.Parameter(b.view(-1), requires_grad=True)
|
||||
|
||||
|
||||
Model = DetectionModel # retain YOLOv5 'Model' class for backwards compatibility
|
||||
|
||||
|
||||
class SegmentationModel(DetectionModel):
|
||||
"""YOLOv5 segmentation model for object detection and segmentation tasks with configurable parameters."""
|
||||
|
||||
def __init__(self, cfg="yolov5s-seg.yaml", ch=3, nc=None, anchors=None):
|
||||
"""Initializes a YOLOv5 segmentation model with configurable params: cfg (str) for configuration, ch (int) for channels, nc (int) for num classes, anchors (list)."""
|
||||
super().__init__(cfg, ch, nc, anchors)
|
||||
|
||||
|
||||
class ClassificationModel(BaseModel):
|
||||
"""YOLOv5 classification model for image classification tasks, initialized with a config file or detection model."""
|
||||
|
||||
def __init__(self, cfg=None, model=None, nc=1000, cutoff=10):
|
||||
"""Initializes YOLOv5 model with config file `cfg`, input channels `ch`, number of classes `nc`, and `cuttoff`
|
||||
index.
|
||||
"""
|
||||
super().__init__()
|
||||
self._from_detection_model(model, nc, cutoff) if model is not None else self._from_yaml(cfg)
|
||||
|
||||
def _from_detection_model(self, model, nc=1000, cutoff=10):
|
||||
"""Creates a classification model from a YOLOv5 detection model, slicing at `cutoff` and adding a classification
|
||||
layer.
|
||||
"""
|
||||
if isinstance(model, DetectMultiBackend):
|
||||
model = model.model # unwrap DetectMultiBackend
|
||||
model.model = model.model[:cutoff] # backbone
|
||||
m = model.model[-1] # last layer
|
||||
ch = m.conv.in_channels if hasattr(m, "conv") else m.cv1.conv.in_channels # ch into module
|
||||
c = Classify(ch, nc) # Classify()
|
||||
c.i, c.f, c.type = m.i, m.f, "models.common.Classify" # index, from, type
|
||||
model.model[-1] = c # replace
|
||||
self.model = model.model
|
||||
self.stride = model.stride
|
||||
self.save = []
|
||||
self.nc = nc
|
||||
|
||||
def _from_yaml(self, cfg):
|
||||
"""Creates a YOLOv5 classification model from a specified *.yaml configuration file."""
|
||||
self.model = None
|
||||
|
||||
|
||||
def parse_model(d, ch):
|
||||
"""Parses a YOLOv5 model from a dict `d`, configuring layers based on input channels `ch` and model architecture."""
|
||||
LOGGER.info(f"\n{'':>3}{'from':>18}{'n':>3}{'params':>10} {'module':<40}{'arguments':<30}")
|
||||
anchors, nc, gd, gw, act, ch_mul = (
|
||||
d["anchors"],
|
||||
d["nc"],
|
||||
d["depth_multiple"],
|
||||
d["width_multiple"],
|
||||
d.get("activation"),
|
||||
d.get("channel_multiple"),
|
||||
)
|
||||
if act:
|
||||
Conv.default_act = eval(act) # redefine default activation, i.e. Conv.default_act = nn.SiLU()
|
||||
LOGGER.info(f"{colorstr('activation:')} {act}") # print
|
||||
if not ch_mul:
|
||||
ch_mul = 8
|
||||
na = (len(anchors[0]) // 2) if isinstance(anchors, list) else anchors # number of anchors
|
||||
no = na * (nc + 5) # number of outputs = anchors * (classes + 5)
|
||||
|
||||
layers, save, c2 = [], [], ch[-1] # layers, savelist, ch out
|
||||
for i, (f, n, m, args) in enumerate(d["backbone"] + d["head"]): # from, number, module, args
|
||||
m = eval(m) if isinstance(m, str) else m # eval strings
|
||||
for j, a in enumerate(args):
|
||||
with contextlib.suppress(NameError):
|
||||
args[j] = eval(a) if isinstance(a, str) else a # eval strings
|
||||
|
||||
n = n_ = max(round(n * gd), 1) if n > 1 else n # depth gain
|
||||
if m in {
|
||||
Conv,
|
||||
GhostConv,
|
||||
Bottleneck,
|
||||
GhostBottleneck,
|
||||
SPP,
|
||||
SPPF,
|
||||
DWConv,
|
||||
MixConv2d,
|
||||
Focus,
|
||||
CrossConv,
|
||||
BottleneckCSP,
|
||||
C3,
|
||||
C3TR,
|
||||
C3SPP,
|
||||
C3Ghost,
|
||||
nn.ConvTranspose2d,
|
||||
DWConvTranspose2d,
|
||||
C3x,
|
||||
}:
|
||||
c1, c2 = ch[f], args[0]
|
||||
if c2 != no: # if not output
|
||||
c2 = make_divisible(c2 * gw, ch_mul)
|
||||
|
||||
args = [c1, c2, *args[1:]]
|
||||
if m in {BottleneckCSP, C3, C3TR, C3Ghost, C3x}:
|
||||
args.insert(2, n) # number of repeats
|
||||
n = 1
|
||||
elif m is nn.BatchNorm2d:
|
||||
args = [ch[f]]
|
||||
elif m is Concat:
|
||||
c2 = sum(ch[x] for x in f)
|
||||
# TODO: channel, gw, gd
|
||||
elif m in {Detect, Segment}:
|
||||
args.append([ch[x] for x in f])
|
||||
if isinstance(args[1], int): # number of anchors
|
||||
args[1] = [list(range(args[1] * 2))] * len(f)
|
||||
if m is Segment:
|
||||
args[3] = make_divisible(args[3] * gw, ch_mul)
|
||||
elif m is Contract:
|
||||
c2 = ch[f] * args[0] ** 2
|
||||
elif m is Expand:
|
||||
c2 = ch[f] // args[0] ** 2
|
||||
else:
|
||||
c2 = ch[f]
|
||||
|
||||
m_ = nn.Sequential(*(m(*args) for _ in range(n))) if n > 1 else m(*args) # module
|
||||
t = str(m)[8:-2].replace("__main__.", "") # module type
|
||||
np = sum(x.numel() for x in m_.parameters()) # number params
|
||||
m_.i, m_.f, m_.type, m_.np = i, f, t, np # attach index, 'from' index, type, number params
|
||||
LOGGER.info(f"{i:>3}{str(f):>18}{n_:>3}{np:10.0f} {t:<40}{str(args):<30}") # print
|
||||
save.extend(x % i for x in ([f] if isinstance(f, int) else f) if x != -1) # append to savelist
|
||||
layers.append(m_)
|
||||
if i == 0:
|
||||
ch = []
|
||||
ch.append(c2)
|
||||
return nn.Sequential(*layers), sorted(save)
|
||||
|
||||
|
||||
if __name__ == "__main__":
|
||||
parser = argparse.ArgumentParser()
|
||||
parser.add_argument("--cfg", type=str, default="yolov5s.yaml", help="model.yaml")
|
||||
parser.add_argument("--batch-size", type=int, default=1, help="total batch size for all GPUs")
|
||||
parser.add_argument("--device", default="", help="cuda device, i.e. 0 or 0,1,2,3 or cpu")
|
||||
parser.add_argument("--profile", action="store_true", help="profile model speed")
|
||||
parser.add_argument("--line-profile", action="store_true", help="profile model speed layer by layer")
|
||||
parser.add_argument("--test", action="store_true", help="test all yolo*.yaml")
|
||||
opt = parser.parse_args()
|
||||
opt.cfg = check_yaml(opt.cfg) # check YAML
|
||||
print_args(vars(opt))
|
||||
device = select_device(opt.device)
|
||||
|
||||
# Create model
|
||||
im = torch.rand(opt.batch_size, 3, 640, 640).to(device)
|
||||
model = Model(opt.cfg).to(device)
|
||||
|
||||
# Options
|
||||
if opt.line_profile: # profile layer by layer
|
||||
model(im, profile=True)
|
||||
|
||||
elif opt.profile: # profile forward-backward
|
||||
results = profile(input=im, ops=[model], n=3)
|
||||
|
||||
elif opt.test: # test all models
|
||||
for cfg in Path(ROOT / "models").rglob("yolo*.yaml"):
|
||||
try:
|
||||
_ = Model(cfg)
|
||||
except Exception as e:
|
||||
print(f"Error in {cfg}: {e}")
|
||||
|
||||
else: # report fused model summary
|
||||
model.fuse()
|
||||
@@ -1,49 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 1.0 # model depth multiple
|
||||
width_multiple: 1.0 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 13
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 20 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [1024, False]], # 23 (P5/32-large)
|
||||
|
||||
[[17, 20, 23], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,49 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 0.67 # model depth multiple
|
||||
width_multiple: 0.75 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 13
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 20 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [1024, False]], # 23 (P5/32-large)
|
||||
|
||||
[[17, 20, 23], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,49 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 0.33 # model depth multiple
|
||||
width_multiple: 0.25 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 13
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 20 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [1024, False]], # 23 (P5/32-large)
|
||||
|
||||
[[17, 20, 23], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,49 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 0.33 # model depth multiple
|
||||
width_multiple: 0.50 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 13
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 20 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [1024, False]], # 23 (P5/32-large)
|
||||
|
||||
[[17, 20, 23], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,49 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Parameters
|
||||
nc: 80 # number of classes
|
||||
depth_multiple: 1.33 # model depth multiple
|
||||
width_multiple: 1.25 # layer channel multiple
|
||||
anchors:
|
||||
- [10, 13, 16, 30, 33, 23] # P3/8
|
||||
- [30, 61, 62, 45, 59, 119] # P4/16
|
||||
- [116, 90, 156, 198, 373, 326] # P5/32
|
||||
|
||||
# YOLOv5 v6.0 backbone
|
||||
backbone:
|
||||
# [from, number, module, args]
|
||||
[
|
||||
[-1, 1, Conv, [64, 6, 2, 2]], # 0-P1/2
|
||||
[-1, 1, Conv, [128, 3, 2]], # 1-P2/4
|
||||
[-1, 3, C3, [128]],
|
||||
[-1, 1, Conv, [256, 3, 2]], # 3-P3/8
|
||||
[-1, 6, C3, [256]],
|
||||
[-1, 1, Conv, [512, 3, 2]], # 5-P4/16
|
||||
[-1, 9, C3, [512]],
|
||||
[-1, 1, Conv, [1024, 3, 2]], # 7-P5/32
|
||||
[-1, 3, C3, [1024]],
|
||||
[-1, 1, SPPF, [1024, 5]], # 9
|
||||
]
|
||||
|
||||
# YOLOv5 v6.0 head
|
||||
head: [
|
||||
[-1, 1, Conv, [512, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 6], 1, Concat, [1]], # cat backbone P4
|
||||
[-1, 3, C3, [512, False]], # 13
|
||||
|
||||
[-1, 1, Conv, [256, 1, 1]],
|
||||
[-1, 1, nn.Upsample, [None, 2, "nearest"]],
|
||||
[[-1, 4], 1, Concat, [1]], # cat backbone P3
|
||||
[-1, 3, C3, [256, False]], # 17 (P3/8-small)
|
||||
|
||||
[-1, 1, Conv, [256, 3, 2]],
|
||||
[[-1, 14], 1, Concat, [1]], # cat head P4
|
||||
[-1, 3, C3, [512, False]], # 20 (P4/16-medium)
|
||||
|
||||
[-1, 1, Conv, [512, 3, 2]],
|
||||
[[-1, 10], 1, Concat, [1]], # cat head P5
|
||||
[-1, 3, C3, [1024, False]], # 23 (P5/32-large)
|
||||
|
||||
[[17, 20, 23], 1, Detect, [nc, anchors]], # Detect(P3, P4, P5)
|
||||
]
|
||||
@@ -1,97 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""utils/initialization."""
|
||||
|
||||
import contextlib
|
||||
import platform
|
||||
import threading
|
||||
|
||||
|
||||
def emojis(str=""):
|
||||
"""Returns an emoji-safe version of a string, stripped of emojis on Windows platforms."""
|
||||
return str.encode().decode("ascii", "ignore") if platform.system() == "Windows" else str
|
||||
|
||||
|
||||
class TryExcept(contextlib.ContextDecorator):
|
||||
"""A context manager and decorator for error handling that prints an optional message with emojis on exception."""
|
||||
|
||||
def __init__(self, msg=""):
|
||||
"""Initializes TryExcept with an optional message, used as a decorator or context manager for error handling."""
|
||||
self.msg = msg
|
||||
|
||||
def __enter__(self):
|
||||
"""Enter the runtime context related to this object for error handling with an optional message."""
|
||||
pass
|
||||
|
||||
def __exit__(self, exc_type, value, traceback):
|
||||
"""Context manager exit method that prints an error message with emojis if an exception occurred, always returns
|
||||
True.
|
||||
"""
|
||||
if value:
|
||||
print(emojis(f"{self.msg}{': ' if self.msg else ''}{value}"))
|
||||
return True
|
||||
|
||||
|
||||
def threaded(func):
|
||||
"""Decorator @threaded to run a function in a separate thread, returning the thread instance."""
|
||||
|
||||
def wrapper(*args, **kwargs):
|
||||
"""Runs the decorated function in a separate daemon thread and returns the thread instance."""
|
||||
thread = threading.Thread(target=func, args=args, kwargs=kwargs, daemon=True)
|
||||
thread.start()
|
||||
return thread
|
||||
|
||||
return wrapper
|
||||
|
||||
|
||||
def join_threads(verbose=False):
|
||||
"""
|
||||
Joins all daemon threads, optionally printing their names if verbose is True.
|
||||
|
||||
Example: atexit.register(lambda: join_threads())
|
||||
"""
|
||||
main_thread = threading.current_thread()
|
||||
for t in threading.enumerate():
|
||||
if t is not main_thread:
|
||||
if verbose:
|
||||
print(f"Joining thread {t.name}")
|
||||
t.join()
|
||||
|
||||
|
||||
def notebook_init(verbose=True):
|
||||
"""Initializes notebook environment by checking requirements, cleaning up, and displaying system info."""
|
||||
print("Checking setup...")
|
||||
|
||||
import os
|
||||
import shutil
|
||||
|
||||
from ultralytics.utils.checks import check_requirements
|
||||
|
||||
from utils.general import check_font, is_colab
|
||||
from utils.torch_utils import select_device # imports
|
||||
|
||||
check_font()
|
||||
|
||||
import psutil
|
||||
|
||||
if check_requirements("wandb", install=False):
|
||||
os.system("pip uninstall -y wandb") # eliminate unexpected account creation prompt with infinite hang
|
||||
if is_colab():
|
||||
shutil.rmtree("/content/sample_data", ignore_errors=True) # remove colab /sample_data directory
|
||||
|
||||
# System info
|
||||
display = None
|
||||
if verbose:
|
||||
gb = 1 << 30 # bytes to GiB (1024 ** 3)
|
||||
ram = psutil.virtual_memory().total
|
||||
total, used, free = shutil.disk_usage("/")
|
||||
with contextlib.suppress(Exception): # clear display if ipython is installed
|
||||
from IPython import display
|
||||
|
||||
display.clear_output()
|
||||
s = f"({os.cpu_count()} CPUs, {ram / gb:.1f} GB RAM, {(total - free) / gb:.1f}/{total / gb:.1f} GB disk)"
|
||||
else:
|
||||
s = ""
|
||||
|
||||
select_device(newline=False)
|
||||
print(emojis(f"Setup complete ✅ {s}"))
|
||||
return display
|
||||
@@ -1,134 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Activation functions."""
|
||||
|
||||
import torch
|
||||
import torch.nn as nn
|
||||
import torch.nn.functional as F
|
||||
|
||||
|
||||
class SiLU(nn.Module):
|
||||
"""Applies the Sigmoid-weighted Linear Unit (SiLU) activation function, also known as Swish."""
|
||||
|
||||
@staticmethod
|
||||
def forward(x):
|
||||
"""
|
||||
Applies the Sigmoid-weighted Linear Unit (SiLU) activation function.
|
||||
|
||||
https://arxiv.org/pdf/1606.08415.pdf.
|
||||
"""
|
||||
return x * torch.sigmoid(x)
|
||||
|
||||
|
||||
class Hardswish(nn.Module):
|
||||
"""Applies the Hardswish activation function, which is efficient for mobile and embedded devices."""
|
||||
|
||||
@staticmethod
|
||||
def forward(x):
|
||||
"""
|
||||
Applies the Hardswish activation function, compatible with TorchScript, CoreML, and ONNX.
|
||||
|
||||
Equivalent to x * F.hardsigmoid(x)
|
||||
"""
|
||||
return x * F.hardtanh(x + 3, 0.0, 6.0) / 6.0 # for TorchScript, CoreML and ONNX
|
||||
|
||||
|
||||
class Mish(nn.Module):
|
||||
"""Mish activation https://github.com/digantamisra98/Mish."""
|
||||
|
||||
@staticmethod
|
||||
def forward(x):
|
||||
"""Applies the Mish activation function, a smooth alternative to ReLU."""
|
||||
return x * F.softplus(x).tanh()
|
||||
|
||||
|
||||
class MemoryEfficientMish(nn.Module):
|
||||
"""Efficiently applies the Mish activation function using custom autograd for reduced memory usage."""
|
||||
|
||||
class F(torch.autograd.Function):
|
||||
"""Implements a custom autograd function for memory-efficient Mish activation."""
|
||||
|
||||
@staticmethod
|
||||
def forward(ctx, x):
|
||||
"""Applies the Mish activation function, a smooth ReLU alternative, to the input tensor `x`."""
|
||||
ctx.save_for_backward(x)
|
||||
return x.mul(torch.tanh(F.softplus(x))) # x * tanh(ln(1 + exp(x)))
|
||||
|
||||
@staticmethod
|
||||
def backward(ctx, grad_output):
|
||||
"""Computes the gradient of the Mish activation function with respect to input `x`."""
|
||||
x = ctx.saved_tensors[0]
|
||||
sx = torch.sigmoid(x)
|
||||
fx = F.softplus(x).tanh()
|
||||
return grad_output * (fx + x * sx * (1 - fx * fx))
|
||||
|
||||
def forward(self, x):
|
||||
"""Applies the Mish activation function to the input tensor `x`."""
|
||||
return self.F.apply(x)
|
||||
|
||||
|
||||
class FReLU(nn.Module):
|
||||
"""FReLU activation https://arxiv.org/abs/2007.11824."""
|
||||
|
||||
def __init__(self, c1, k=3): # ch_in, kernel
|
||||
"""Initializes FReLU activation with channel `c1` and kernel size `k`."""
|
||||
super().__init__()
|
||||
self.conv = nn.Conv2d(c1, c1, k, 1, 1, groups=c1, bias=False)
|
||||
self.bn = nn.BatchNorm2d(c1)
|
||||
|
||||
def forward(self, x):
|
||||
"""
|
||||
Applies FReLU activation with max operation between input and BN-convolved input.
|
||||
|
||||
https://arxiv.org/abs/2007.11824
|
||||
"""
|
||||
return torch.max(x, self.bn(self.conv(x)))
|
||||
|
||||
|
||||
class AconC(nn.Module):
|
||||
"""
|
||||
ACON activation (activate or not) function.
|
||||
|
||||
AconC: (p1*x-p2*x) * sigmoid(beta*(p1*x-p2*x)) + p2*x, beta is a learnable parameter
|
||||
See "Activate or Not: Learning Customized Activation" https://arxiv.org/pdf/2009.04759.pdf.
|
||||
"""
|
||||
|
||||
def __init__(self, c1):
|
||||
"""Initializes AconC with learnable parameters p1, p2, and beta for channel-wise activation control."""
|
||||
super().__init__()
|
||||
self.p1 = nn.Parameter(torch.randn(1, c1, 1, 1))
|
||||
self.p2 = nn.Parameter(torch.randn(1, c1, 1, 1))
|
||||
self.beta = nn.Parameter(torch.ones(1, c1, 1, 1))
|
||||
|
||||
def forward(self, x):
|
||||
"""Applies AconC activation function with learnable parameters for channel-wise control on input tensor x."""
|
||||
dpx = (self.p1 - self.p2) * x
|
||||
return dpx * torch.sigmoid(self.beta * dpx) + self.p2 * x
|
||||
|
||||
|
||||
class MetaAconC(nn.Module):
|
||||
"""
|
||||
ACON activation (activate or not) function.
|
||||
|
||||
AconC: (p1*x-p2*x) * sigmoid(beta*(p1*x-p2*x)) + p2*x, beta is a learnable parameter
|
||||
See "Activate or Not: Learning Customized Activation" https://arxiv.org/pdf/2009.04759.pdf.
|
||||
"""
|
||||
|
||||
def __init__(self, c1, k=1, s=1, r=16):
|
||||
"""Initializes MetaAconC with params: channel_in (c1), kernel size (k=1), stride (s=1), reduction (r=16)."""
|
||||
super().__init__()
|
||||
c2 = max(r, c1 // r)
|
||||
self.p1 = nn.Parameter(torch.randn(1, c1, 1, 1))
|
||||
self.p2 = nn.Parameter(torch.randn(1, c1, 1, 1))
|
||||
self.fc1 = nn.Conv2d(c1, c2, k, s, bias=True)
|
||||
self.fc2 = nn.Conv2d(c2, c1, k, s, bias=True)
|
||||
# self.bn1 = nn.BatchNorm2d(c2)
|
||||
# self.bn2 = nn.BatchNorm2d(c1)
|
||||
|
||||
def forward(self, x):
|
||||
"""Applies a forward pass transforming input `x` using learnable parameters and sigmoid activation."""
|
||||
y = x.mean(dim=2, keepdims=True).mean(dim=3, keepdims=True)
|
||||
# batch-size 1 bug/instabilities https://github.com/ultralytics/yolov5/issues/2891
|
||||
# beta = torch.sigmoid(self.bn2(self.fc2(self.bn1(self.fc1(y))))) # bug/unstable
|
||||
beta = torch.sigmoid(self.fc2(self.fc1(y))) # bug patch BN layers removed
|
||||
dpx = (self.p1 - self.p2) * x
|
||||
return dpx * torch.sigmoid(beta * dpx) + self.p2 * x
|
||||
@@ -1,440 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Image augmentation functions."""
|
||||
|
||||
import math
|
||||
import random
|
||||
|
||||
import cv2
|
||||
import numpy as np
|
||||
import torch
|
||||
import torchvision.transforms as T
|
||||
import torchvision.transforms.functional as TF
|
||||
|
||||
from utils.yolov5.utils.general import LOGGER, check_version, colorstr, resample_segments, segment2box, xywhn2xyxy
|
||||
from utils.yolov5.utils.metrics import bbox_ioa
|
||||
|
||||
IMAGENET_MEAN = 0.485, 0.456, 0.406 # RGB mean
|
||||
IMAGENET_STD = 0.229, 0.224, 0.225 # RGB standard deviation
|
||||
|
||||
|
||||
class Albumentations:
|
||||
"""Provides optional data augmentation for YOLOv5 using Albumentations library if installed."""
|
||||
|
||||
def __init__(self, size=640):
|
||||
"""Initializes Albumentations class for optional data augmentation in YOLOv5 with specified input size."""
|
||||
self.transform = None
|
||||
prefix = colorstr("albumentations: ")
|
||||
try:
|
||||
import albumentations as A
|
||||
|
||||
check_version(A.__version__, "1.0.3", hard=True) # version requirement
|
||||
|
||||
T = [
|
||||
A.RandomResizedCrop(height=size, width=size, scale=(0.8, 1.0), ratio=(0.9, 1.11), p=0.0),
|
||||
A.Blur(p=0.01),
|
||||
A.MedianBlur(p=0.01),
|
||||
A.ToGray(p=0.01),
|
||||
A.CLAHE(p=0.01),
|
||||
A.RandomBrightnessContrast(p=0.0),
|
||||
A.RandomGamma(p=0.0),
|
||||
A.ImageCompression(quality_lower=75, p=0.0),
|
||||
] # transforms
|
||||
self.transform = A.Compose(T, bbox_params=A.BboxParams(format="yolo", label_fields=["class_labels"]))
|
||||
|
||||
LOGGER.info(prefix + ", ".join(f"{x}".replace("always_apply=False, ", "") for x in T if x.p))
|
||||
except ImportError: # package not installed, skip
|
||||
pass
|
||||
except Exception as e:
|
||||
LOGGER.info(f"{prefix}{e}")
|
||||
|
||||
def __call__(self, im, labels, p=1.0):
|
||||
"""Applies transformations to an image and labels with probability `p`, returning updated image and labels."""
|
||||
if self.transform and random.random() < p:
|
||||
new = self.transform(image=im, bboxes=labels[:, 1:], class_labels=labels[:, 0]) # transformed
|
||||
im, labels = new["image"], np.array([[c, *b] for c, b in zip(new["class_labels"], new["bboxes"])])
|
||||
return im, labels
|
||||
|
||||
|
||||
def normalize(x, mean=IMAGENET_MEAN, std=IMAGENET_STD, inplace=False):
|
||||
"""
|
||||
Applies ImageNet normalization to RGB images in BCHW format, modifying them in-place if specified.
|
||||
|
||||
Example: y = (x - mean) / std
|
||||
"""
|
||||
return TF.normalize(x, mean, std, inplace=inplace)
|
||||
|
||||
|
||||
def denormalize(x, mean=IMAGENET_MEAN, std=IMAGENET_STD):
|
||||
"""Reverses ImageNet normalization for BCHW format RGB images by applying `x = x * std + mean`."""
|
||||
for i in range(3):
|
||||
x[:, i] = x[:, i] * std[i] + mean[i]
|
||||
return x
|
||||
|
||||
|
||||
def augment_hsv(im, hgain=0.5, sgain=0.5, vgain=0.5):
|
||||
"""Applies HSV color-space augmentation to an image with random gains for hue, saturation, and value."""
|
||||
if hgain or sgain or vgain:
|
||||
r = np.random.uniform(-1, 1, 3) * [hgain, sgain, vgain] + 1 # random gains
|
||||
hue, sat, val = cv2.split(cv2.cvtColor(im, cv2.COLOR_BGR2HSV))
|
||||
dtype = im.dtype # uint8
|
||||
|
||||
x = np.arange(0, 256, dtype=r.dtype)
|
||||
lut_hue = ((x * r[0]) % 180).astype(dtype)
|
||||
lut_sat = np.clip(x * r[1], 0, 255).astype(dtype)
|
||||
lut_val = np.clip(x * r[2], 0, 255).astype(dtype)
|
||||
|
||||
im_hsv = cv2.merge((cv2.LUT(hue, lut_hue), cv2.LUT(sat, lut_sat), cv2.LUT(val, lut_val)))
|
||||
cv2.cvtColor(im_hsv, cv2.COLOR_HSV2BGR, dst=im) # no return needed
|
||||
|
||||
|
||||
def hist_equalize(im, clahe=True, bgr=False):
|
||||
"""Equalizes image histogram, with optional CLAHE, for BGR or RGB image with shape (n,m,3) and range 0-255."""
|
||||
yuv = cv2.cvtColor(im, cv2.COLOR_BGR2YUV if bgr else cv2.COLOR_RGB2YUV)
|
||||
if clahe:
|
||||
c = cv2.createCLAHE(clipLimit=2.0, tileGridSize=(8, 8))
|
||||
yuv[:, :, 0] = c.apply(yuv[:, :, 0])
|
||||
else:
|
||||
yuv[:, :, 0] = cv2.equalizeHist(yuv[:, :, 0]) # equalize Y channel histogram
|
||||
return cv2.cvtColor(yuv, cv2.COLOR_YUV2BGR if bgr else cv2.COLOR_YUV2RGB) # convert YUV image to RGB
|
||||
|
||||
|
||||
def replicate(im, labels):
|
||||
"""
|
||||
Replicates half of the smallest object labels in an image for data augmentation.
|
||||
|
||||
Returns augmented image and labels.
|
||||
"""
|
||||
h, w = im.shape[:2]
|
||||
boxes = labels[:, 1:].astype(int)
|
||||
x1, y1, x2, y2 = boxes.T
|
||||
s = ((x2 - x1) + (y2 - y1)) / 2 # side length (pixels)
|
||||
for i in s.argsort()[: round(s.size * 0.5)]: # smallest indices
|
||||
x1b, y1b, x2b, y2b = boxes[i]
|
||||
bh, bw = y2b - y1b, x2b - x1b
|
||||
yc, xc = int(random.uniform(0, h - bh)), int(random.uniform(0, w - bw)) # offset x, y
|
||||
x1a, y1a, x2a, y2a = [xc, yc, xc + bw, yc + bh]
|
||||
im[y1a:y2a, x1a:x2a] = im[y1b:y2b, x1b:x2b] # im4[ymin:ymax, xmin:xmax]
|
||||
labels = np.append(labels, [[labels[i, 0], x1a, y1a, x2a, y2a]], axis=0)
|
||||
|
||||
return im, labels
|
||||
|
||||
|
||||
def letterbox(im, new_shape=(640, 640), color=(114, 114, 114), auto=True, scaleFill=False, scaleup=True, stride=32):
|
||||
"""Resizes and pads image to new_shape with stride-multiple constraints, returns resized image, ratio, padding."""
|
||||
shape = im.shape[:2] # current shape [height, width]
|
||||
if isinstance(new_shape, int):
|
||||
new_shape = (new_shape, new_shape)
|
||||
|
||||
# Scale ratio (new / old)
|
||||
r = min(new_shape[0] / shape[0], new_shape[1] / shape[1])
|
||||
if not scaleup: # only scale down, do not scale up (for better val mAP)
|
||||
r = min(r, 1.0)
|
||||
|
||||
# Compute padding
|
||||
ratio = r, r # width, height ratios
|
||||
new_unpad = int(round(shape[1] * r)), int(round(shape[0] * r))
|
||||
dw, dh = new_shape[1] - new_unpad[0], new_shape[0] - new_unpad[1] # wh padding
|
||||
if auto: # minimum rectangle
|
||||
dw, dh = np.mod(dw, stride), np.mod(dh, stride) # wh padding
|
||||
elif scaleFill: # stretch
|
||||
dw, dh = 0.0, 0.0
|
||||
new_unpad = (new_shape[1], new_shape[0])
|
||||
ratio = new_shape[1] / shape[1], new_shape[0] / shape[0] # width, height ratios
|
||||
|
||||
dw /= 2 # divide padding into 2 sides
|
||||
dh /= 2
|
||||
|
||||
if shape[::-1] != new_unpad: # resize
|
||||
im = cv2.resize(im, new_unpad, interpolation=cv2.INTER_LINEAR)
|
||||
top, bottom = int(round(dh - 0.1)), int(round(dh + 0.1))
|
||||
left, right = int(round(dw - 0.1)), int(round(dw + 0.1))
|
||||
im = cv2.copyMakeBorder(im, top, bottom, left, right, cv2.BORDER_CONSTANT, value=color) # add border
|
||||
return im, ratio, (dw, dh)
|
||||
|
||||
|
||||
def random_perspective(
|
||||
im, targets=(), segments=(), degrees=10, translate=0.1, scale=0.1, shear=10, perspective=0.0, border=(0, 0)
|
||||
):
|
||||
# torchvision.transforms.RandomAffine(degrees=(-10, 10), translate=(0.1, 0.1), scale=(0.9, 1.1), shear=(-10, 10))
|
||||
# targets = [cls, xyxy]
|
||||
"""Applies random perspective transformation to an image, modifying the image and corresponding labels."""
|
||||
height = im.shape[0] + border[0] * 2 # shape(h,w,c)
|
||||
width = im.shape[1] + border[1] * 2
|
||||
|
||||
# Center
|
||||
C = np.eye(3)
|
||||
C[0, 2] = -im.shape[1] / 2 # x translation (pixels)
|
||||
C[1, 2] = -im.shape[0] / 2 # y translation (pixels)
|
||||
|
||||
# Perspective
|
||||
P = np.eye(3)
|
||||
P[2, 0] = random.uniform(-perspective, perspective) # x perspective (about y)
|
||||
P[2, 1] = random.uniform(-perspective, perspective) # y perspective (about x)
|
||||
|
||||
# Rotation and Scale
|
||||
R = np.eye(3)
|
||||
a = random.uniform(-degrees, degrees)
|
||||
# a += random.choice([-180, -90, 0, 90]) # add 90deg rotations to small rotations
|
||||
s = random.uniform(1 - scale, 1 + scale)
|
||||
# s = 2 ** random.uniform(-scale, scale)
|
||||
R[:2] = cv2.getRotationMatrix2D(angle=a, center=(0, 0), scale=s)
|
||||
|
||||
# Shear
|
||||
S = np.eye(3)
|
||||
S[0, 1] = math.tan(random.uniform(-shear, shear) * math.pi / 180) # x shear (deg)
|
||||
S[1, 0] = math.tan(random.uniform(-shear, shear) * math.pi / 180) # y shear (deg)
|
||||
|
||||
# Translation
|
||||
T = np.eye(3)
|
||||
T[0, 2] = random.uniform(0.5 - translate, 0.5 + translate) * width # x translation (pixels)
|
||||
T[1, 2] = random.uniform(0.5 - translate, 0.5 + translate) * height # y translation (pixels)
|
||||
|
||||
# Combined rotation matrix
|
||||
M = T @ S @ R @ P @ C # order of operations (right to left) is IMPORTANT
|
||||
if (border[0] != 0) or (border[1] != 0) or (M != np.eye(3)).any(): # image changed
|
||||
if perspective:
|
||||
im = cv2.warpPerspective(im, M, dsize=(width, height), borderValue=(114, 114, 114))
|
||||
else: # affine
|
||||
im = cv2.warpAffine(im, M[:2], dsize=(width, height), borderValue=(114, 114, 114))
|
||||
|
||||
if n := len(targets):
|
||||
use_segments = any(x.any() for x in segments) and len(segments) == n
|
||||
new = np.zeros((n, 4))
|
||||
if use_segments: # warp segments
|
||||
segments = resample_segments(segments) # upsample
|
||||
for i, segment in enumerate(segments):
|
||||
xy = np.ones((len(segment), 3))
|
||||
xy[:, :2] = segment
|
||||
xy = xy @ M.T # transform
|
||||
xy = xy[:, :2] / xy[:, 2:3] if perspective else xy[:, :2] # perspective rescale or affine
|
||||
|
||||
# clip
|
||||
new[i] = segment2box(xy, width, height)
|
||||
|
||||
else: # warp boxes
|
||||
xy = np.ones((n * 4, 3))
|
||||
xy[:, :2] = targets[:, [1, 2, 3, 4, 1, 4, 3, 2]].reshape(n * 4, 2) # x1y1, x2y2, x1y2, x2y1
|
||||
xy = xy @ M.T # transform
|
||||
xy = (xy[:, :2] / xy[:, 2:3] if perspective else xy[:, :2]).reshape(n, 8) # perspective rescale or affine
|
||||
|
||||
# create new boxes
|
||||
x = xy[:, [0, 2, 4, 6]]
|
||||
y = xy[:, [1, 3, 5, 7]]
|
||||
new = np.concatenate((x.min(1), y.min(1), x.max(1), y.max(1))).reshape(4, n).T
|
||||
|
||||
# clip
|
||||
new[:, [0, 2]] = new[:, [0, 2]].clip(0, width)
|
||||
new[:, [1, 3]] = new[:, [1, 3]].clip(0, height)
|
||||
|
||||
# filter candidates
|
||||
i = box_candidates(box1=targets[:, 1:5].T * s, box2=new.T, area_thr=0.01 if use_segments else 0.10)
|
||||
targets = targets[i]
|
||||
targets[:, 1:5] = new[i]
|
||||
|
||||
return im, targets
|
||||
|
||||
|
||||
def copy_paste(im, labels, segments, p=0.5):
|
||||
"""
|
||||
Applies Copy-Paste augmentation by flipping and merging segments and labels on an image.
|
||||
|
||||
Details at https://arxiv.org/abs/2012.07177.
|
||||
"""
|
||||
n = len(segments)
|
||||
if p and n:
|
||||
h, w, c = im.shape # height, width, channels
|
||||
im_new = np.zeros(im.shape, np.uint8)
|
||||
for j in random.sample(range(n), k=round(p * n)):
|
||||
l, s = labels[j], segments[j]
|
||||
box = w - l[3], l[2], w - l[1], l[4]
|
||||
ioa = bbox_ioa(box, labels[:, 1:5]) # intersection over area
|
||||
if (ioa < 0.30).all(): # allow 30% obscuration of existing labels
|
||||
labels = np.concatenate((labels, [[l[0], *box]]), 0)
|
||||
segments.append(np.concatenate((w - s[:, 0:1], s[:, 1:2]), 1))
|
||||
cv2.drawContours(im_new, [segments[j].astype(np.int32)], -1, (1, 1, 1), cv2.FILLED)
|
||||
|
||||
result = cv2.flip(im, 1) # augment segments (flip left-right)
|
||||
i = cv2.flip(im_new, 1).astype(bool)
|
||||
im[i] = result[i] # cv2.imwrite('debug.jpg', im) # debug
|
||||
|
||||
return im, labels, segments
|
||||
|
||||
|
||||
def cutout(im, labels, p=0.5):
|
||||
"""
|
||||
Applies cutout augmentation to an image with optional label adjustment, using random masks of varying sizes.
|
||||
|
||||
Details at https://arxiv.org/abs/1708.04552.
|
||||
"""
|
||||
if random.random() < p:
|
||||
h, w = im.shape[:2]
|
||||
scales = [0.5] * 1 + [0.25] * 2 + [0.125] * 4 + [0.0625] * 8 + [0.03125] * 16 # image size fraction
|
||||
for s in scales:
|
||||
mask_h = random.randint(1, int(h * s)) # create random masks
|
||||
mask_w = random.randint(1, int(w * s))
|
||||
|
||||
# box
|
||||
xmin = max(0, random.randint(0, w) - mask_w // 2)
|
||||
ymin = max(0, random.randint(0, h) - mask_h // 2)
|
||||
xmax = min(w, xmin + mask_w)
|
||||
ymax = min(h, ymin + mask_h)
|
||||
|
||||
# apply random color mask
|
||||
im[ymin:ymax, xmin:xmax] = [random.randint(64, 191) for _ in range(3)]
|
||||
|
||||
# return unobscured labels
|
||||
if len(labels) and s > 0.03:
|
||||
box = np.array([xmin, ymin, xmax, ymax], dtype=np.float32)
|
||||
ioa = bbox_ioa(box, xywhn2xyxy(labels[:, 1:5], w, h)) # intersection over area
|
||||
labels = labels[ioa < 0.60] # remove >60% obscured labels
|
||||
|
||||
return labels
|
||||
|
||||
|
||||
def mixup(im, labels, im2, labels2):
|
||||
"""
|
||||
Applies MixUp augmentation by blending images and labels.
|
||||
|
||||
See https://arxiv.org/pdf/1710.09412.pdf for details.
|
||||
"""
|
||||
r = np.random.beta(32.0, 32.0) # mixup ratio, alpha=beta=32.0
|
||||
im = (im * r + im2 * (1 - r)).astype(np.uint8)
|
||||
labels = np.concatenate((labels, labels2), 0)
|
||||
return im, labels
|
||||
|
||||
|
||||
def box_candidates(box1, box2, wh_thr=2, ar_thr=100, area_thr=0.1, eps=1e-16):
|
||||
"""
|
||||
Filters bounding box candidates by minimum width-height threshold `wh_thr` (pixels), aspect ratio threshold
|
||||
`ar_thr`, and area ratio threshold `area_thr`.
|
||||
|
||||
box1(4,n) is before augmentation, box2(4,n) is after augmentation.
|
||||
"""
|
||||
w1, h1 = box1[2] - box1[0], box1[3] - box1[1]
|
||||
w2, h2 = box2[2] - box2[0], box2[3] - box2[1]
|
||||
ar = np.maximum(w2 / (h2 + eps), h2 / (w2 + eps)) # aspect ratio
|
||||
return (w2 > wh_thr) & (h2 > wh_thr) & (w2 * h2 / (w1 * h1 + eps) > area_thr) & (ar < ar_thr) # candidates
|
||||
|
||||
|
||||
def classify_albumentations(
|
||||
augment=True,
|
||||
size=224,
|
||||
scale=(0.08, 1.0),
|
||||
ratio=(0.75, 1.0 / 0.75), # 0.75, 1.33
|
||||
hflip=0.5,
|
||||
vflip=0.0,
|
||||
jitter=0.4,
|
||||
mean=IMAGENET_MEAN,
|
||||
std=IMAGENET_STD,
|
||||
auto_aug=False,
|
||||
):
|
||||
# YOLOv5 classification Albumentations (optional, only used if package is installed)
|
||||
"""Sets up and returns Albumentations transforms for YOLOv5 classification tasks depending on augmentation
|
||||
settings.
|
||||
"""
|
||||
prefix = colorstr("albumentations: ")
|
||||
try:
|
||||
import albumentations as A
|
||||
from albumentations.pytorch import ToTensorV2
|
||||
|
||||
check_version(A.__version__, "1.0.3", hard=True) # version requirement
|
||||
if augment: # Resize and crop
|
||||
T = [A.RandomResizedCrop(height=size, width=size, scale=scale, ratio=ratio)]
|
||||
if auto_aug:
|
||||
# TODO: implement AugMix, AutoAug & RandAug in albumentation
|
||||
LOGGER.info(f"{prefix}auto augmentations are currently not supported")
|
||||
else:
|
||||
if hflip > 0:
|
||||
T += [A.HorizontalFlip(p=hflip)]
|
||||
if vflip > 0:
|
||||
T += [A.VerticalFlip(p=vflip)]
|
||||
if jitter > 0:
|
||||
color_jitter = (float(jitter),) * 3 # repeat value for brightness, contrast, saturation, 0 hue
|
||||
T += [A.ColorJitter(*color_jitter, 0)]
|
||||
else: # Use fixed crop for eval set (reproducibility)
|
||||
T = [A.SmallestMaxSize(max_size=size), A.CenterCrop(height=size, width=size)]
|
||||
T += [A.Normalize(mean=mean, std=std), ToTensorV2()] # Normalize and convert to Tensor
|
||||
LOGGER.info(prefix + ", ".join(f"{x}".replace("always_apply=False, ", "") for x in T if x.p))
|
||||
return A.Compose(T)
|
||||
|
||||
except ImportError: # package not installed, skip
|
||||
LOGGER.warning(f"{prefix}⚠️ not found, install with `pip install albumentations` (recommended)")
|
||||
except Exception as e:
|
||||
LOGGER.info(f"{prefix}{e}")
|
||||
|
||||
|
||||
def classify_transforms(size=224):
|
||||
"""Applies a series of transformations including center crop, ToTensor, and normalization for classification."""
|
||||
assert isinstance(size, int), f"ERROR: classify_transforms size {size} must be integer, not (list, tuple)"
|
||||
# T.Compose([T.ToTensor(), T.Resize(size), T.CenterCrop(size), T.Normalize(IMAGENET_MEAN, IMAGENET_STD)])
|
||||
return T.Compose([CenterCrop(size), ToTensor(), T.Normalize(IMAGENET_MEAN, IMAGENET_STD)])
|
||||
|
||||
|
||||
class LetterBox:
|
||||
"""Resizes and pads images to specified dimensions while maintaining aspect ratio for YOLOv5 preprocessing."""
|
||||
|
||||
def __init__(self, size=(640, 640), auto=False, stride=32):
|
||||
"""Initializes a LetterBox object for YOLOv5 image preprocessing with optional auto sizing and stride
|
||||
adjustment.
|
||||
"""
|
||||
super().__init__()
|
||||
self.h, self.w = (size, size) if isinstance(size, int) else size
|
||||
self.auto = auto # pass max size integer, automatically solve for short side using stride
|
||||
self.stride = stride # used with auto
|
||||
|
||||
def __call__(self, im):
|
||||
"""
|
||||
Resizes and pads input image `im` (HWC format) to specified dimensions, maintaining aspect ratio.
|
||||
|
||||
im = np.array HWC
|
||||
"""
|
||||
imh, imw = im.shape[:2]
|
||||
r = min(self.h / imh, self.w / imw) # ratio of new/old
|
||||
h, w = round(imh * r), round(imw * r) # resized image
|
||||
hs, ws = (math.ceil(x / self.stride) * self.stride for x in (h, w)) if self.auto else self.h, self.w
|
||||
top, left = round((hs - h) / 2 - 0.1), round((ws - w) / 2 - 0.1)
|
||||
im_out = np.full((self.h, self.w, 3), 114, dtype=im.dtype)
|
||||
im_out[top : top + h, left : left + w] = cv2.resize(im, (w, h), interpolation=cv2.INTER_LINEAR)
|
||||
return im_out
|
||||
|
||||
|
||||
class CenterCrop:
|
||||
"""Applies center crop to an image, resizing it to the specified size while maintaining aspect ratio."""
|
||||
|
||||
def __init__(self, size=640):
|
||||
"""Initializes CenterCrop for image preprocessing, accepting single int or tuple for size, defaults to 640."""
|
||||
super().__init__()
|
||||
self.h, self.w = (size, size) if isinstance(size, int) else size
|
||||
|
||||
def __call__(self, im):
|
||||
"""
|
||||
Applies center crop to the input image and resizes it to a specified size, maintaining aspect ratio.
|
||||
|
||||
im = np.array HWC
|
||||
"""
|
||||
imh, imw = im.shape[:2]
|
||||
m = min(imh, imw) # min dimension
|
||||
top, left = (imh - m) // 2, (imw - m) // 2
|
||||
return cv2.resize(im[top : top + m, left : left + m], (self.w, self.h), interpolation=cv2.INTER_LINEAR)
|
||||
|
||||
|
||||
class ToTensor:
|
||||
"""Converts BGR np.array image from HWC to RGB CHW format, normalizes to [0, 1], and supports FP16 if half=True."""
|
||||
|
||||
def __init__(self, half=False):
|
||||
"""Initializes ToTensor for YOLOv5 image preprocessing, with optional half precision (half=True for FP16)."""
|
||||
super().__init__()
|
||||
self.half = half
|
||||
|
||||
def __call__(self, im):
|
||||
"""
|
||||
Converts BGR np.array image from HWC to RGB CHW format, and normalizes to [0, 1], with support for FP16 if
|
||||
`half=True`.
|
||||
|
||||
im = np.array HWC in BGR order
|
||||
"""
|
||||
im = np.ascontiguousarray(im.transpose((2, 0, 1))[::-1]) # HWC to CHW -> BGR to RGB -> contiguous
|
||||
im = torch.from_numpy(im) # to torch
|
||||
im = im.half() if self.half else im.float() # uint8 to fp16/32
|
||||
im /= 255.0 # 0-255 to 0.0-1.0
|
||||
return im
|
||||
@@ -1,175 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""AutoAnchor utils."""
|
||||
|
||||
import random
|
||||
|
||||
import numpy as np
|
||||
import torch
|
||||
import yaml
|
||||
from tqdm import tqdm
|
||||
|
||||
from utils.yolov5.utils import TryExcept
|
||||
from utils.yolov5.utils.general import LOGGER, TQDM_BAR_FORMAT, colorstr
|
||||
|
||||
PREFIX = colorstr("AutoAnchor: ")
|
||||
|
||||
|
||||
def check_anchor_order(m):
|
||||
"""Checks and corrects anchor order against stride in YOLOv5 Detect() module if necessary."""
|
||||
a = m.anchors.prod(-1).mean(-1).view(-1) # mean anchor area per output layer
|
||||
da = a[-1] - a[0] # delta a
|
||||
ds = m.stride[-1] - m.stride[0] # delta s
|
||||
if da and (da.sign() != ds.sign()): # same order
|
||||
LOGGER.info(f"{PREFIX}Reversing anchor order")
|
||||
m.anchors[:] = m.anchors.flip(0)
|
||||
|
||||
|
||||
@TryExcept(f"{PREFIX}ERROR")
|
||||
def check_anchors(dataset, model, thr=4.0, imgsz=640):
|
||||
"""Evaluates anchor fit to dataset and adjusts if necessary, supporting customizable threshold and image size."""
|
||||
m = model.module.model[-1] if hasattr(model, "module") else model.model[-1] # Detect()
|
||||
shapes = imgsz * dataset.shapes / dataset.shapes.max(1, keepdims=True)
|
||||
scale = np.random.uniform(0.9, 1.1, size=(shapes.shape[0], 1)) # augment scale
|
||||
wh = torch.tensor(np.concatenate([l[:, 3:5] * s for s, l in zip(shapes * scale, dataset.labels)])).float() # wh
|
||||
|
||||
def metric(k): # compute metric
|
||||
"""Computes ratio metric, anchors above threshold, and best possible recall for YOLOv5 anchor evaluation."""
|
||||
r = wh[:, None] / k[None]
|
||||
x = torch.min(r, 1 / r).min(2)[0] # ratio metric
|
||||
best = x.max(1)[0] # best_x
|
||||
aat = (x > 1 / thr).float().sum(1).mean() # anchors above threshold
|
||||
bpr = (best > 1 / thr).float().mean() # best possible recall
|
||||
return bpr, aat
|
||||
|
||||
stride = m.stride.to(m.anchors.device).view(-1, 1, 1) # model strides
|
||||
anchors = m.anchors.clone() * stride # current anchors
|
||||
bpr, aat = metric(anchors.cpu().view(-1, 2))
|
||||
s = f"\n{PREFIX}{aat:.2f} anchors/target, {bpr:.3f} Best Possible Recall (BPR). "
|
||||
if bpr > 0.98: # threshold to recompute
|
||||
LOGGER.info(f"{s}Current anchors are a good fit to dataset ✅")
|
||||
else:
|
||||
LOGGER.info(f"{s}Anchors are a poor fit to dataset ⚠️, attempting to improve...")
|
||||
na = m.anchors.numel() // 2 # number of anchors
|
||||
anchors = kmean_anchors(dataset, n=na, img_size=imgsz, thr=thr, gen=1000, verbose=False)
|
||||
new_bpr = metric(anchors)[0]
|
||||
if new_bpr > bpr: # replace anchors
|
||||
anchors = torch.tensor(anchors, device=m.anchors.device).type_as(m.anchors)
|
||||
m.anchors[:] = anchors.clone().view_as(m.anchors)
|
||||
check_anchor_order(m) # must be in pixel-space (not grid-space)
|
||||
m.anchors /= stride
|
||||
s = f"{PREFIX}Done ✅ (optional: update model *.yaml to use these anchors in the future)"
|
||||
else:
|
||||
s = f"{PREFIX}Done ⚠️ (original anchors better than new anchors, proceeding with original anchors)"
|
||||
LOGGER.info(s)
|
||||
|
||||
|
||||
def kmean_anchors(dataset="./data/coco128.yaml", n=9, img_size=640, thr=4.0, gen=1000, verbose=True):
|
||||
"""
|
||||
Creates kmeans-evolved anchors from training dataset.
|
||||
|
||||
Arguments:
|
||||
dataset: path to data.yaml, or a loaded dataset
|
||||
n: number of anchors
|
||||
img_size: image size used for training
|
||||
thr: anchor-label wh ratio threshold hyperparameter hyp['anchor_t'] used for training, default=4.0
|
||||
gen: generations to evolve anchors using genetic algorithm
|
||||
verbose: print all results
|
||||
|
||||
Return:
|
||||
k: kmeans evolved anchors
|
||||
|
||||
Usage:
|
||||
from utils.autoanchor import *; _ = kmean_anchors()
|
||||
"""
|
||||
from scipy.cluster.vq import kmeans
|
||||
|
||||
npr = np.random
|
||||
thr = 1 / thr
|
||||
|
||||
def metric(k, wh): # compute metrics
|
||||
"""Computes ratio metric, anchors above threshold, and best possible recall for YOLOv5 anchor evaluation."""
|
||||
r = wh[:, None] / k[None]
|
||||
x = torch.min(r, 1 / r).min(2)[0] # ratio metric
|
||||
# x = wh_iou(wh, torch.tensor(k)) # iou metric
|
||||
return x, x.max(1)[0] # x, best_x
|
||||
|
||||
def anchor_fitness(k): # mutation fitness
|
||||
"""Evaluates fitness of YOLOv5 anchors by computing recall and ratio metrics for an anchor evolution process."""
|
||||
_, best = metric(torch.tensor(k, dtype=torch.float32), wh)
|
||||
return (best * (best > thr).float()).mean() # fitness
|
||||
|
||||
def print_results(k, verbose=True):
|
||||
"""Sorts and logs kmeans-evolved anchor metrics and best possible recall values for YOLOv5 anchor evaluation."""
|
||||
k = k[np.argsort(k.prod(1))] # sort small to large
|
||||
x, best = metric(k, wh0)
|
||||
bpr, aat = (best > thr).float().mean(), (x > thr).float().mean() * n # best possible recall, anch > thr
|
||||
s = (
|
||||
f"{PREFIX}thr={thr:.2f}: {bpr:.4f} best possible recall, {aat:.2f} anchors past thr\n"
|
||||
f"{PREFIX}n={n}, img_size={img_size}, metric_all={x.mean():.3f}/{best.mean():.3f}-mean/best, "
|
||||
f"past_thr={x[x > thr].mean():.3f}-mean: "
|
||||
)
|
||||
for x in k:
|
||||
s += "%i,%i, " % (round(x[0]), round(x[1]))
|
||||
if verbose:
|
||||
LOGGER.info(s[:-2])
|
||||
return k
|
||||
|
||||
if isinstance(dataset, str): # *.yaml file
|
||||
with open(dataset, errors="ignore") as f:
|
||||
data_dict = yaml.safe_load(f) # model dict
|
||||
from utils.dataloaders import LoadImagesAndLabels
|
||||
|
||||
dataset = LoadImagesAndLabels(data_dict["train"], augment=True, rect=True)
|
||||
|
||||
# Get label wh
|
||||
shapes = img_size * dataset.shapes / dataset.shapes.max(1, keepdims=True)
|
||||
wh0 = np.concatenate([l[:, 3:5] * s for s, l in zip(shapes, dataset.labels)]) # wh
|
||||
|
||||
# Filter
|
||||
i = (wh0 < 3.0).any(1).sum()
|
||||
if i:
|
||||
LOGGER.info(f"{PREFIX}WARNING ⚠️ Extremely small objects found: {i} of {len(wh0)} labels are <3 pixels in size")
|
||||
wh = wh0[(wh0 >= 2.0).any(1)].astype(np.float32) # filter > 2 pixels
|
||||
# wh = wh * (npr.rand(wh.shape[0], 1) * 0.9 + 0.1) # multiply by random scale 0-1
|
||||
|
||||
# Kmeans init
|
||||
try:
|
||||
LOGGER.info(f"{PREFIX}Running kmeans for {n} anchors on {len(wh)} points...")
|
||||
assert n <= len(wh) # apply overdetermined constraint
|
||||
s = wh.std(0) # sigmas for whitening
|
||||
k = kmeans(wh / s, n, iter=30)[0] * s # points
|
||||
assert n == len(k) # kmeans may return fewer points than requested if wh is insufficient or too similar
|
||||
except Exception:
|
||||
LOGGER.warning(f"{PREFIX}WARNING ⚠️ switching strategies from kmeans to random init")
|
||||
k = np.sort(npr.rand(n * 2)).reshape(n, 2) * img_size # random init
|
||||
wh, wh0 = (torch.tensor(x, dtype=torch.float32) for x in (wh, wh0))
|
||||
k = print_results(k, verbose=False)
|
||||
|
||||
# Plot
|
||||
# k, d = [None] * 20, [None] * 20
|
||||
# for i in tqdm(range(1, 21)):
|
||||
# k[i-1], d[i-1] = kmeans(wh / s, i) # points, mean distance
|
||||
# fig, ax = plt.subplots(1, 2, figsize=(14, 7), tight_layout=True)
|
||||
# ax = ax.ravel()
|
||||
# ax[0].plot(np.arange(1, 21), np.array(d) ** 2, marker='.')
|
||||
# fig, ax = plt.subplots(1, 2, figsize=(14, 7)) # plot wh
|
||||
# ax[0].hist(wh[wh[:, 0]<100, 0],400)
|
||||
# ax[1].hist(wh[wh[:, 1]<100, 1],400)
|
||||
# fig.savefig('wh.png', dpi=200)
|
||||
|
||||
# Evolve
|
||||
f, sh, mp, s = anchor_fitness(k), k.shape, 0.9, 0.1 # fitness, generations, mutation prob, sigma
|
||||
pbar = tqdm(range(gen), bar_format=TQDM_BAR_FORMAT) # progress bar
|
||||
for _ in pbar:
|
||||
v = np.ones(sh)
|
||||
while (v == 1).all(): # mutate until a change occurs (prevent duplicates)
|
||||
v = ((npr.random(sh) < mp) * random.random() * npr.randn(*sh) * s + 1).clip(0.3, 3.0)
|
||||
kg = (k.copy() * v).clip(min=2.0)
|
||||
fg = anchor_fitness(kg)
|
||||
if fg > f:
|
||||
f, k = fg, kg.copy()
|
||||
pbar.desc = f"{PREFIX}Evolving anchors with Genetic Algorithm: fitness = {f:.4f}"
|
||||
if verbose:
|
||||
print_results(k, verbose)
|
||||
|
||||
return print_results(k).astype(np.float32)
|
||||
@@ -1,70 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Auto-batch utils."""
|
||||
|
||||
from copy import deepcopy
|
||||
|
||||
import numpy as np
|
||||
import torch
|
||||
|
||||
from utils.yolov5.utils.general import LOGGER, colorstr
|
||||
from utils.yolov5.utils.torch_utils import profile
|
||||
|
||||
|
||||
def check_train_batch_size(model, imgsz=640, amp=True):
|
||||
"""Checks and computes optimal training batch size for YOLOv5 model, given image size and AMP setting."""
|
||||
with torch.cuda.amp.autocast(amp):
|
||||
return autobatch(deepcopy(model).train(), imgsz) # compute optimal batch size
|
||||
|
||||
|
||||
def autobatch(model, imgsz=640, fraction=0.8, batch_size=16):
|
||||
"""Estimates optimal YOLOv5 batch size using `fraction` of CUDA memory."""
|
||||
# Usage:
|
||||
# import torch
|
||||
# from utils.autobatch import autobatch
|
||||
# model = torch.hub.load('ultralytics/yolov5', 'yolov5s', autoshape=False)
|
||||
# print(autobatch(model))
|
||||
|
||||
# Check device
|
||||
prefix = colorstr("AutoBatch: ")
|
||||
LOGGER.info(f"{prefix}Computing optimal batch size for --imgsz {imgsz}")
|
||||
device = next(model.parameters()).device # get model device
|
||||
if device.type == "cpu":
|
||||
LOGGER.info(f"{prefix}CUDA not detected, using default CPU batch-size {batch_size}")
|
||||
return batch_size
|
||||
if torch.backends.cudnn.benchmark:
|
||||
LOGGER.info(f"{prefix} ⚠️ Requires torch.backends.cudnn.benchmark=False, using default batch-size {batch_size}")
|
||||
return batch_size
|
||||
|
||||
# Inspect CUDA memory
|
||||
gb = 1 << 30 # bytes to GiB (1024 ** 3)
|
||||
d = str(device).upper() # 'CUDA:0'
|
||||
properties = torch.cuda.get_device_properties(device) # device properties
|
||||
t = properties.total_memory / gb # GiB total
|
||||
r = torch.cuda.memory_reserved(device) / gb # GiB reserved
|
||||
a = torch.cuda.memory_allocated(device) / gb # GiB allocated
|
||||
f = t - (r + a) # GiB free
|
||||
LOGGER.info(f"{prefix}{d} ({properties.name}) {t:.2f}G total, {r:.2f}G reserved, {a:.2f}G allocated, {f:.2f}G free")
|
||||
|
||||
# Profile batch sizes
|
||||
batch_sizes = [1, 2, 4, 8, 16]
|
||||
try:
|
||||
img = [torch.empty(b, 3, imgsz, imgsz) for b in batch_sizes]
|
||||
results = profile(img, model, n=3, device=device)
|
||||
except Exception as e:
|
||||
LOGGER.warning(f"{prefix}{e}")
|
||||
|
||||
# Fit a solution
|
||||
y = [x[2] for x in results if x] # memory [2]
|
||||
p = np.polyfit(batch_sizes[: len(y)], y, deg=1) # first degree polynomial fit
|
||||
b = int((f * fraction - p[1]) / p[0]) # y intercept (optimal batch size)
|
||||
if None in results: # some sizes failed
|
||||
i = results.index(None) # first fail index
|
||||
if b >= batch_sizes[i]: # y intercept above failure point
|
||||
b = batch_sizes[max(i - 1, 0)] # select prior safe point
|
||||
if b < 1 or b > 1024: # b outside of safe range
|
||||
b = batch_size
|
||||
LOGGER.warning(f"{prefix}WARNING ⚠️ CUDA anomaly detected, recommend restart environment and retry command.")
|
||||
|
||||
fraction = (np.polyval(p, b) + r + a) / t # actual fraction predicted
|
||||
LOGGER.info(f"{prefix}Using batch-size {b} for {d} {t * fraction:.2f}G/{t:.2f}G ({fraction * 100:.0f}%) ✅")
|
||||
return b
|
||||
@@ -1 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
@@ -1,26 +0,0 @@
|
||||
# AWS EC2 instance startup 'MIME' script https://aws.amazon.com/premiumsupport/knowledge-center/execute-user-data-ec2/
|
||||
# This script will run on every instance restart, not only on first start
|
||||
# --- DO NOT COPY ABOVE COMMENTS WHEN PASTING INTO USERDATA ---
|
||||
|
||||
Content-Type: multipart/mixed; boundary="//"
|
||||
MIME-Version: 1.0
|
||||
|
||||
--//
|
||||
Content-Type: text/cloud-config; charset="us-ascii"
|
||||
MIME-Version: 1.0
|
||||
Content-Transfer-Encoding: 7bit
|
||||
Content-Disposition: attachment; filename="cloud-config.txt"
|
||||
|
||||
#cloud-config
|
||||
cloud_final_modules:
|
||||
- [scripts-user, always]
|
||||
|
||||
--//
|
||||
Content-Type: text/x-shellscript; charset="us-ascii"
|
||||
MIME-Version: 1.0
|
||||
Content-Transfer-Encoding: 7bit
|
||||
Content-Disposition: attachment; filename="userdata.txt"
|
||||
|
||||
#!/bin/bash
|
||||
# --- paste contents of userdata.sh here ---
|
||||
--//
|
||||
@@ -1,42 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# Resume all interrupted trainings in yolov5/ dir including DDP trainings
|
||||
# Usage: $ python utils/aws/resume.py
|
||||
|
||||
import os
|
||||
import sys
|
||||
from pathlib import Path
|
||||
|
||||
import torch
|
||||
import yaml
|
||||
|
||||
FILE = Path(__file__).resolve()
|
||||
ROOT = FILE.parents[2] # YOLOv5 root directory
|
||||
if str(ROOT) not in sys.path:
|
||||
sys.path.append(str(ROOT)) # add ROOT to PATH
|
||||
|
||||
port = 0 # --master_port
|
||||
path = Path("").resolve()
|
||||
for last in path.rglob("*/**/last.pt"):
|
||||
ckpt = torch.load(last)
|
||||
if ckpt["optimizer"] is None:
|
||||
continue
|
||||
|
||||
# Load opt.yaml
|
||||
with open(last.parent.parent / "opt.yaml", errors="ignore") as f:
|
||||
opt = yaml.safe_load(f)
|
||||
|
||||
# Get device count
|
||||
d = opt["device"].split(",") # devices
|
||||
nd = len(d) # number of devices
|
||||
ddp = nd > 1 or (nd == 0 and torch.cuda.device_count() > 1) # distributed data parallel
|
||||
|
||||
if ddp: # multi-GPU
|
||||
port += 1
|
||||
cmd = f"python -m torch.distributed.run --nproc_per_node {nd} --master_port {port} train.py --resume {last}"
|
||||
else: # single-GPU
|
||||
cmd = f"python train.py --resume {last}"
|
||||
|
||||
cmd += " > /dev/null 2>&1 &" # redirect output to dev/null and run in daemon thread
|
||||
print(cmd)
|
||||
os.system(cmd)
|
||||
@@ -1,27 +0,0 @@
|
||||
#!/bin/bash
|
||||
# AWS EC2 instance startup script https://docs.aws.amazon.com/AWSEC2/latest/UserGuide/user-data.html
|
||||
# This script will run only once on first instance start (for a re-start script see mime.sh)
|
||||
# /home/ubuntu (ubuntu) or /home/ec2-user (amazon-linux) is working dir
|
||||
# Use >300 GB SSD
|
||||
|
||||
cd home/ubuntu
|
||||
if [ ! -d yolov5 ]; then
|
||||
echo "Running first-time script." # install dependencies, download COCO, pull Docker
|
||||
git clone https://github.com/ultralytics/yolov5 -b master && sudo chmod -R 777 yolov5
|
||||
cd yolov5
|
||||
bash data/scripts/get_coco.sh && echo "COCO done." &
|
||||
sudo docker pull ultralytics/yolov5:latest && echo "Docker done." &
|
||||
python -m pip install --upgrade pip && pip install -r requirements.txt && python detect.py && echo "Requirements done." &
|
||||
wait && echo "All tasks done." # finish background tasks
|
||||
else
|
||||
echo "Running re-start script." # resume interrupted runs
|
||||
i=0
|
||||
list=$(sudo docker ps -qa) # container list i.e. $'one\ntwo\nthree\nfour'
|
||||
while IFS= read -r id; do
|
||||
((i++))
|
||||
echo "restarting container $i: $id"
|
||||
sudo docker start $id
|
||||
# sudo docker exec -it $id python train.py --resume # single-GPU
|
||||
sudo docker exec -d $id python utils/aws/resume.py # multi-scenario
|
||||
done <<<"$list"
|
||||
fi
|
||||
@@ -1,72 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Callback utils."""
|
||||
|
||||
import threading
|
||||
|
||||
|
||||
class Callbacks:
|
||||
"""Handles all registered callbacks for YOLOv5 Hooks."""
|
||||
|
||||
def __init__(self):
|
||||
"""Initializes a Callbacks object to manage registered YOLOv5 training event hooks."""
|
||||
self._callbacks = {
|
||||
"on_pretrain_routine_start": [],
|
||||
"on_pretrain_routine_end": [],
|
||||
"on_train_start": [],
|
||||
"on_train_epoch_start": [],
|
||||
"on_train_batch_start": [],
|
||||
"optimizer_step": [],
|
||||
"on_before_zero_grad": [],
|
||||
"on_train_batch_end": [],
|
||||
"on_train_epoch_end": [],
|
||||
"on_val_start": [],
|
||||
"on_val_batch_start": [],
|
||||
"on_val_image_end": [],
|
||||
"on_val_batch_end": [],
|
||||
"on_val_end": [],
|
||||
"on_fit_epoch_end": [], # fit = train + val
|
||||
"on_model_save": [],
|
||||
"on_train_end": [],
|
||||
"on_params_update": [],
|
||||
"teardown": [],
|
||||
}
|
||||
self.stop_training = False # set True to interrupt training
|
||||
|
||||
def register_action(self, hook, name="", callback=None):
|
||||
"""
|
||||
Register a new action to a callback hook.
|
||||
|
||||
Args:
|
||||
hook: The callback hook name to register the action to
|
||||
name: The name of the action for later reference
|
||||
callback: The callback to fire
|
||||
"""
|
||||
assert hook in self._callbacks, f"hook '{hook}' not found in callbacks {self._callbacks}"
|
||||
assert callable(callback), f"callback '{callback}' is not callable"
|
||||
self._callbacks[hook].append({"name": name, "callback": callback})
|
||||
|
||||
def get_registered_actions(self, hook=None):
|
||||
"""
|
||||
Returns all the registered actions by callback hook.
|
||||
|
||||
Args:
|
||||
hook: The name of the hook to check, defaults to all
|
||||
"""
|
||||
return self._callbacks[hook] if hook else self._callbacks
|
||||
|
||||
def run(self, hook, *args, thread=False, **kwargs):
|
||||
"""
|
||||
Loop through the registered actions and fire all callbacks on main thread.
|
||||
|
||||
Args:
|
||||
hook: The name of the hook to check, defaults to all
|
||||
args: Arguments to receive from YOLOv5
|
||||
thread: (boolean) Run callbacks in daemon thread
|
||||
kwargs: Keyword Arguments to receive from YOLOv5
|
||||
"""
|
||||
assert hook in self._callbacks, f"hook '{hook}' not found in callbacks {self._callbacks}"
|
||||
for logger in self._callbacks[hook]:
|
||||
if thread:
|
||||
threading.Thread(target=logger["callback"], args=args, kwargs=kwargs, daemon=True).start()
|
||||
else:
|
||||
logger["callback"](*args, **kwargs)
|
||||
File diff suppressed because it is too large
Load Diff
@@ -1,73 +0,0 @@
|
||||
# YOLOv5 🚀 by Ultralytics, AGPL-3.0 license
|
||||
# Builds ultralytics/yolov5:latest image on DockerHub https://hub.docker.com/r/ultralytics/yolov5
|
||||
# Image is CUDA-optimized for YOLOv5 single/multi-GPU training and inference
|
||||
|
||||
# Start FROM PyTorch image https://hub.docker.com/r/pytorch/pytorch
|
||||
FROM pytorch/pytorch:2.0.0-cuda11.7-cudnn8-runtime
|
||||
|
||||
# Downloads to user config dir
|
||||
ADD https://ultralytics.com/assets/Arial.ttf https://ultralytics.com/assets/Arial.Unicode.ttf /root/.config/Ultralytics/
|
||||
|
||||
# Install linux packages
|
||||
ENV DEBIAN_FRONTEND noninteractive
|
||||
RUN apt update
|
||||
RUN TZ=Etc/UTC apt install -y tzdata
|
||||
RUN apt install --no-install-recommends -y gcc git zip curl htop libgl1 libglib2.0-0 libpython3-dev gnupg
|
||||
# RUN alias python=python3
|
||||
|
||||
# Security updates
|
||||
# https://security.snyk.io/vuln/SNYK-UBUNTU1804-OPENSSL-3314796
|
||||
RUN apt upgrade --no-install-recommends -y openssl
|
||||
|
||||
# Create working directory
|
||||
RUN rm -rf /usr/src/app && mkdir -p /usr/src/app
|
||||
WORKDIR /usr/src/app
|
||||
|
||||
# Copy contents
|
||||
COPY . /usr/src/app
|
||||
|
||||
# Install pip packages
|
||||
COPY requirements.txt .
|
||||
RUN python3 -m pip install --upgrade pip wheel
|
||||
RUN pip install --no-cache -r requirements.txt albumentations comet gsutil notebook \
|
||||
coremltools onnx onnx-simplifier onnxruntime 'openvino-dev>=2023.0'
|
||||
# tensorflow tensorflowjs \
|
||||
|
||||
# Set environment variables
|
||||
ENV OMP_NUM_THREADS=1
|
||||
|
||||
# Cleanup
|
||||
ENV DEBIAN_FRONTEND teletype
|
||||
|
||||
|
||||
# Usage Examples -------------------------------------------------------------------------------------------------------
|
||||
|
||||
# Build and Push
|
||||
# t=ultralytics/yolov5:latest && sudo docker build -f utils/docker/Dockerfile -t $t . && sudo docker push $t
|
||||
|
||||
# Pull and Run
|
||||
# t=ultralytics/yolov5:latest && sudo docker pull $t && sudo docker run -it --ipc=host --gpus all $t
|
||||
|
||||
# Pull and Run with local directory access
|
||||
# t=ultralytics/yolov5:latest && sudo docker pull $t && sudo docker run -it --ipc=host --gpus all -v "$(pwd)"/datasets:/usr/src/datasets $t
|
||||
|
||||
# Kill all
|
||||
# sudo docker kill $(sudo docker ps -q)
|
||||
|
||||
# Kill all image-based
|
||||
# sudo docker kill $(sudo docker ps -qa --filter ancestor=ultralytics/yolov5:latest)
|
||||
|
||||
# DockerHub tag update
|
||||
# t=ultralytics/yolov5:latest tnew=ultralytics/yolov5:v6.2 && sudo docker pull $t && sudo docker tag $t $tnew && sudo docker push $tnew
|
||||
|
||||
# Clean up
|
||||
# sudo docker system prune -a --volumes
|
||||
|
||||
# Update Ubuntu drivers
|
||||
# https://www.maketecheasier.com/install-nvidia-drivers-ubuntu/
|
||||
|
||||
# DDP test
|
||||
# python -m torch.distributed.run --nproc_per_node 2 --master_port 1 train.py --epochs 3
|
||||
|
||||
# GCP VM from Image
|
||||
# docker.io/ultralytics/yolov5:latest
|
||||
@@ -1,40 +0,0 @@
|
||||
# YOLOv5 🚀 by Ultralytics, AGPL-3.0 license
|
||||
# Builds ultralytics/yolov5:latest-arm64 image on DockerHub https://hub.docker.com/r/ultralytics/yolov5
|
||||
# Image is aarch64-compatible for Apple M1 and other ARM architectures i.e. Jetson Nano and Raspberry Pi
|
||||
|
||||
# Start FROM Ubuntu image https://hub.docker.com/_/ubuntu
|
||||
FROM arm64v8/ubuntu:22.10
|
||||
|
||||
# Downloads to user config dir
|
||||
ADD https://ultralytics.com/assets/Arial.ttf https://ultralytics.com/assets/Arial.Unicode.ttf /root/.config/Ultralytics/
|
||||
|
||||
# Install linux packages
|
||||
ENV DEBIAN_FRONTEND noninteractive
|
||||
RUN apt update
|
||||
RUN TZ=Etc/UTC apt install -y tzdata
|
||||
RUN apt install --no-install-recommends -y python3-pip git zip curl htop gcc libgl1 libglib2.0-0 libpython3-dev
|
||||
# RUN alias python=python3
|
||||
|
||||
# Install pip packages
|
||||
COPY requirements.txt .
|
||||
RUN python3 -m pip install --upgrade pip wheel
|
||||
RUN pip install --no-cache -r requirements.txt albumentations gsutil notebook \
|
||||
coremltools onnx onnxruntime
|
||||
# tensorflow-aarch64 tensorflowjs \
|
||||
|
||||
# Create working directory
|
||||
RUN mkdir -p /usr/src/app
|
||||
WORKDIR /usr/src/app
|
||||
|
||||
# Copy contents
|
||||
COPY . /usr/src/app
|
||||
ENV DEBIAN_FRONTEND teletype
|
||||
|
||||
|
||||
# Usage Examples -------------------------------------------------------------------------------------------------------
|
||||
|
||||
# Build and Push
|
||||
# t=ultralytics/yolov5:latest-arm64 && sudo docker build --platform linux/arm64 -f utils/docker/Dockerfile-arm64 -t $t . && sudo docker push $t
|
||||
|
||||
# Pull and Run
|
||||
# t=ultralytics/yolov5:latest-arm64 && sudo docker pull $t && sudo docker run -it --ipc=host -v "$(pwd)"/datasets:/usr/src/datasets $t
|
||||
@@ -1,42 +0,0 @@
|
||||
# YOLOv5 🚀 by Ultralytics, AGPL-3.0 license
|
||||
# Builds ultralytics/yolov5:latest-cpu image on DockerHub https://hub.docker.com/r/ultralytics/yolov5
|
||||
# Image is CPU-optimized for ONNX, OpenVINO and PyTorch YOLOv5 deployments
|
||||
|
||||
# Start FROM Ubuntu image https://hub.docker.com/_/ubuntu
|
||||
FROM ubuntu:23.10
|
||||
|
||||
# Downloads to user config dir
|
||||
ADD https://ultralytics.com/assets/Arial.ttf https://ultralytics.com/assets/Arial.Unicode.ttf /root/.config/Ultralytics/
|
||||
|
||||
# Install linux packages
|
||||
# g++ required to build 'tflite_support' and 'lap' packages, libusb-1.0-0 required for 'tflite_support' package
|
||||
RUN apt update \
|
||||
&& apt install --no-install-recommends -y python3-pip git zip curl htop libgl1 libglib2.0-0 libpython3-dev gnupg g++ libusb-1.0-0
|
||||
# RUN alias python=python3
|
||||
|
||||
# Remove python3.11/EXTERNALLY-MANAGED or use 'pip install --break-system-packages' avoid 'externally-managed-environment' Ubuntu nightly error
|
||||
RUN rm -rf /usr/lib/python3.11/EXTERNALLY-MANAGED
|
||||
|
||||
# Install pip packages
|
||||
COPY requirements.txt .
|
||||
RUN python3 -m pip install --upgrade pip wheel
|
||||
RUN pip install --no-cache -r requirements.txt albumentations gsutil notebook \
|
||||
coremltools onnx onnx-simplifier onnxruntime 'openvino-dev>=2023.0' \
|
||||
# tensorflow tensorflowjs \
|
||||
--extra-index-url https://download.pytorch.org/whl/cpu
|
||||
|
||||
# Create working directory
|
||||
RUN mkdir -p /usr/src/app
|
||||
WORKDIR /usr/src/app
|
||||
|
||||
# Copy contents
|
||||
COPY . /usr/src/app
|
||||
|
||||
|
||||
# Usage Examples -------------------------------------------------------------------------------------------------------
|
||||
|
||||
# Build and Push
|
||||
# t=ultralytics/yolov5:latest-cpu && sudo docker build -f utils/docker/Dockerfile-cpu -t $t . && sudo docker push $t
|
||||
|
||||
# Pull and Run
|
||||
# t=ultralytics/yolov5:latest-cpu && sudo docker pull $t && sudo docker run -it --ipc=host -v "$(pwd)"/datasets:/usr/src/datasets $t
|
||||
@@ -1,136 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Download utils."""
|
||||
|
||||
import logging
|
||||
import subprocess
|
||||
import urllib
|
||||
from pathlib import Path
|
||||
|
||||
import requests
|
||||
import torch
|
||||
|
||||
|
||||
def is_url(url, check=True):
|
||||
"""Determines if a string is a URL and optionally checks its existence online, returning a boolean."""
|
||||
try:
|
||||
url = str(url)
|
||||
result = urllib.parse.urlparse(url)
|
||||
assert all([result.scheme, result.netloc]) # check if is url
|
||||
return (urllib.request.urlopen(url).getcode() == 200) if check else True # check if exists online
|
||||
except (AssertionError, urllib.request.HTTPError):
|
||||
return False
|
||||
|
||||
|
||||
def gsutil_getsize(url=""):
|
||||
"""
|
||||
Returns the size in bytes of a file at a Google Cloud Storage URL using `gsutil du`.
|
||||
|
||||
Returns 0 if the command fails or output is empty.
|
||||
"""
|
||||
output = subprocess.check_output(["gsutil", "du", url], shell=True, encoding="utf-8")
|
||||
return int(output.split()[0]) if output else 0
|
||||
|
||||
|
||||
def url_getsize(url="https://ultralytics.com/images/bus.jpg"):
|
||||
"""Returns the size in bytes of a downloadable file at a given URL; defaults to -1 if not found."""
|
||||
response = requests.head(url, allow_redirects=True)
|
||||
return int(response.headers.get("content-length", -1))
|
||||
|
||||
|
||||
def curl_download(url, filename, *, silent: bool = False) -> bool:
|
||||
"""Download a file from a url to a filename using curl."""
|
||||
silent_option = "sS" if silent else "" # silent
|
||||
proc = subprocess.run(
|
||||
[
|
||||
"curl",
|
||||
"-#",
|
||||
f"-{silent_option}L",
|
||||
url,
|
||||
"--output",
|
||||
filename,
|
||||
"--retry",
|
||||
"9",
|
||||
"-C",
|
||||
"-",
|
||||
]
|
||||
)
|
||||
return proc.returncode == 0
|
||||
|
||||
|
||||
def safe_download(file, url, url2=None, min_bytes=1e0, error_msg=""):
|
||||
"""
|
||||
Downloads a file from a URL (or alternate URL) to a specified path if file is above a minimum size.
|
||||
|
||||
Removes incomplete downloads.
|
||||
"""
|
||||
from utils.yolov5.utils.general import LOGGER
|
||||
|
||||
file = Path(file)
|
||||
assert_msg = f"Downloaded file '{file}' does not exist or size is < min_bytes={min_bytes}"
|
||||
try: # url1
|
||||
LOGGER.info(f"Downloading {url} to {file}...")
|
||||
torch.hub.download_url_to_file(url, str(file), progress=LOGGER.level <= logging.INFO)
|
||||
assert file.exists() and file.stat().st_size > min_bytes, assert_msg # check
|
||||
except Exception as e: # url2
|
||||
if file.exists():
|
||||
file.unlink() # remove partial downloads
|
||||
LOGGER.info(f"ERROR: {e}\nRe-attempting {url2 or url} to {file}...")
|
||||
# curl download, retry and resume on fail
|
||||
curl_download(url2 or url, file)
|
||||
finally:
|
||||
if not file.exists() or file.stat().st_size < min_bytes: # check
|
||||
if file.exists():
|
||||
file.unlink() # remove partial downloads
|
||||
LOGGER.info(f"ERROR: {assert_msg}\n{error_msg}")
|
||||
LOGGER.info("")
|
||||
|
||||
|
||||
def attempt_download(file, repo="ultralytics/yolov5", release="v7.0"):
|
||||
"""Downloads a file from GitHub release assets or via direct URL if not found locally, supporting backup
|
||||
versions.
|
||||
"""
|
||||
from utils.yolov5.utils.general import LOGGER
|
||||
|
||||
def github_assets(repository, version="latest"):
|
||||
"""Fetches GitHub repository release tag and asset names using the GitHub API."""
|
||||
if version != "latest":
|
||||
version = f"tags/{version}" # i.e. tags/v7.0
|
||||
response = requests.get(f"https://api.github.com/repos/{repository}/releases/{version}").json() # github api
|
||||
return response["tag_name"], [x["name"] for x in response["assets"]] # tag, assets
|
||||
|
||||
file = Path(str(file).strip().replace("'", ""))
|
||||
if not file.exists():
|
||||
# URL specified
|
||||
name = Path(urllib.parse.unquote(str(file))).name # decode '%2F' to '/' etc.
|
||||
if str(file).startswith(("http:/", "https:/")): # download
|
||||
url = str(file).replace(":/", "://") # Pathlib turns :// -> :/
|
||||
file = name.split("?")[0] # parse authentication https://url.com/file.txt?auth...
|
||||
if Path(file).is_file():
|
||||
LOGGER.info(f"Found {url} locally at {file}") # file already exists
|
||||
else:
|
||||
safe_download(file=file, url=url, min_bytes=1e5)
|
||||
return file
|
||||
|
||||
# GitHub assets
|
||||
assets = [f"yolov5{size}{suffix}.pt" for size in "nsmlx" for suffix in ("", "6", "-cls", "-seg")] # default
|
||||
try:
|
||||
tag, assets = github_assets(repo, release)
|
||||
except Exception:
|
||||
try:
|
||||
tag, assets = github_assets(repo) # latest release
|
||||
except Exception:
|
||||
try:
|
||||
tag = subprocess.check_output("git tag", shell=True, stderr=subprocess.STDOUT).decode().split()[-1]
|
||||
except Exception:
|
||||
tag = release
|
||||
|
||||
if name in assets:
|
||||
file.parent.mkdir(parents=True, exist_ok=True) # make parent dir (if required)
|
||||
safe_download(
|
||||
file,
|
||||
url=f"https://github.com/{repo}/releases/download/{tag}/{name}",
|
||||
min_bytes=1e5,
|
||||
error_msg=f"{file} missing, try downloading from https://github.com/{repo}/releases/{tag}",
|
||||
)
|
||||
|
||||
return str(file)
|
||||
@@ -1,70 +0,0 @@
|
||||
# Flask REST API
|
||||
|
||||
[REST](https://en.wikipedia.org/wiki/Representational_state_transfer) [API](https://en.wikipedia.org/wiki/API)s are commonly used to expose Machine Learning (ML) models to other services. This folder contains an example REST API created using Flask to expose the YOLOv5s model from [PyTorch Hub](https://pytorch.org/hub/ultralytics_yolov5/).
|
||||
|
||||
## Requirements
|
||||
|
||||
[Flask](https://palletsprojects.com/projects/flask/) is required. Install with:
|
||||
|
||||
```shell
|
||||
$ pip install Flask
|
||||
```
|
||||
|
||||
## Run
|
||||
|
||||
After Flask installation run:
|
||||
|
||||
```shell
|
||||
$ python3 restapi.py --port 5000
|
||||
```
|
||||
|
||||
Then use [curl](https://curl.se/) to perform a request:
|
||||
|
||||
```shell
|
||||
$ curl -X POST -F image=@zidane.jpg 'http://localhost:5000/v1/object-detection/yolov5s'
|
||||
```
|
||||
|
||||
The model inference results are returned as a JSON response:
|
||||
|
||||
```json
|
||||
[
|
||||
{
|
||||
"class": 0,
|
||||
"confidence": 0.8900438547,
|
||||
"height": 0.9318675399,
|
||||
"name": "person",
|
||||
"width": 0.3264600933,
|
||||
"xcenter": 0.7438579798,
|
||||
"ycenter": 0.5207948685
|
||||
},
|
||||
{
|
||||
"class": 0,
|
||||
"confidence": 0.8440024257,
|
||||
"height": 0.7155083418,
|
||||
"name": "person",
|
||||
"width": 0.6546785235,
|
||||
"xcenter": 0.427829951,
|
||||
"ycenter": 0.6334488392
|
||||
},
|
||||
{
|
||||
"class": 27,
|
||||
"confidence": 0.3771208823,
|
||||
"height": 0.3902671337,
|
||||
"name": "tie",
|
||||
"width": 0.0696444362,
|
||||
"xcenter": 0.3675483763,
|
||||
"ycenter": 0.7991207838
|
||||
},
|
||||
{
|
||||
"class": 27,
|
||||
"confidence": 0.3527112305,
|
||||
"height": 0.1540903747,
|
||||
"name": "tie",
|
||||
"width": 0.0336618312,
|
||||
"xcenter": 0.7814827561,
|
||||
"ycenter": 0.5065554976
|
||||
}
|
||||
]
|
||||
```
|
||||
|
||||
An example python script to perform inference using [requests](https://docs.python-requests.org/en/master/) is given in `example_request.py`
|
||||
@@ -1,17 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Perform test request."""
|
||||
|
||||
import pprint
|
||||
|
||||
import requests
|
||||
|
||||
DETECTION_URL = "http://localhost:5000/v1/object-detection/yolov5s"
|
||||
IMAGE = "zidane.jpg"
|
||||
|
||||
# Read image
|
||||
with open(IMAGE, "rb") as f:
|
||||
image_data = f.read()
|
||||
|
||||
response = requests.post(DETECTION_URL, files={"image": image_data}).json()
|
||||
|
||||
pprint.pprint(response)
|
||||
@@ -1,49 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Run a Flask REST API exposing one or more YOLOv5s models."""
|
||||
|
||||
import argparse
|
||||
import io
|
||||
|
||||
import torch
|
||||
from flask import Flask, request
|
||||
from PIL import Image
|
||||
|
||||
app = Flask(__name__)
|
||||
models = {}
|
||||
|
||||
DETECTION_URL = "/v1/object-detection/<model>"
|
||||
|
||||
|
||||
@app.route(DETECTION_URL, methods=["POST"])
|
||||
def predict(model):
|
||||
"""Predict and return object detections in JSON format given an image and model name via a Flask REST API POST
|
||||
request.
|
||||
"""
|
||||
if request.method != "POST":
|
||||
return
|
||||
|
||||
if request.files.get("image"):
|
||||
# Method 1
|
||||
# with request.files["image"] as f:
|
||||
# im = Image.open(io.BytesIO(f.read()))
|
||||
|
||||
# Method 2
|
||||
im_file = request.files["image"]
|
||||
im_bytes = im_file.read()
|
||||
im = Image.open(io.BytesIO(im_bytes))
|
||||
|
||||
if model in models:
|
||||
results = models[model](im, size=640) # reduce size=320 for faster inference
|
||||
return results.pandas().xyxy[0].to_json(orient="records")
|
||||
|
||||
|
||||
if __name__ == "__main__":
|
||||
parser = argparse.ArgumentParser(description="Flask API exposing YOLOv5 model")
|
||||
parser.add_argument("--port", default=5000, type=int, help="port number")
|
||||
parser.add_argument("--model", nargs="+", default=["yolov5s"], help="model(s) to run, i.e. --model yolov5n yolov5s")
|
||||
opt = parser.parse_args()
|
||||
|
||||
for m in opt.model:
|
||||
models[m] = torch.hub.load("ultralytics/yolov5", m, force_reload=True, skip_validation=True)
|
||||
|
||||
app.run(host="0.0.0.0", port=opt.port) # debug=True causes Restarting with stat
|
||||
File diff suppressed because it is too large
Load Diff
@@ -1,25 +0,0 @@
|
||||
FROM gcr.io/google-appengine/python
|
||||
|
||||
# Create a virtualenv for dependencies. This isolates these packages from
|
||||
# system-level packages.
|
||||
# Use -p python3 or -p python3.7 to select python version. Default is version 2.
|
||||
RUN virtualenv /env -p python3
|
||||
|
||||
# Setting these environment variables are the same as running
|
||||
# source /env/bin/activate.
|
||||
ENV VIRTUAL_ENV /env
|
||||
ENV PATH /env/bin:$PATH
|
||||
|
||||
RUN apt-get update && apt-get install -y python-opencv
|
||||
|
||||
# Copy the application's requirements.txt and run pip to install all
|
||||
# dependencies into the virtualenv.
|
||||
ADD requirements.txt /app/requirements.txt
|
||||
RUN pip install -r /app/requirements.txt
|
||||
|
||||
# Add the application source code.
|
||||
ADD . /app
|
||||
|
||||
# Run a WSGI server to serve the application. gunicorn must be declared as
|
||||
# a dependency in requirements.txt.
|
||||
CMD gunicorn -b :$PORT main:app
|
||||
@@ -1,6 +0,0 @@
|
||||
# add these requirements in your app on top of the existing ones
|
||||
pip==23.3
|
||||
Flask==2.3.2
|
||||
gunicorn==22.0.0
|
||||
werkzeug>=3.0.1 # not directly required, pinned by Snyk to avoid a vulnerability
|
||||
zipp>=3.19.1 # not directly required, pinned by Snyk to avoid a vulnerability
|
||||
@@ -1,16 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
runtime: custom
|
||||
env: flex
|
||||
|
||||
service: yolov5app
|
||||
|
||||
liveness_check:
|
||||
initial_delay_sec: 600
|
||||
|
||||
manual_scaling:
|
||||
instances: 1
|
||||
resources:
|
||||
cpu: 1
|
||||
memory_gb: 4
|
||||
disk_size_gb: 20
|
||||
@@ -1,476 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Logging utils."""
|
||||
|
||||
import json
|
||||
import os
|
||||
import warnings
|
||||
from pathlib import Path
|
||||
|
||||
import pkg_resources as pkg
|
||||
import torch
|
||||
|
||||
from utils.general import LOGGER, colorstr, cv2
|
||||
from utils.loggers.clearml.clearml_utils import ClearmlLogger
|
||||
from utils.loggers.wandb.wandb_utils import WandbLogger
|
||||
from utils.plots import plot_images, plot_labels, plot_results
|
||||
from utils.torch_utils import de_parallel
|
||||
|
||||
LOGGERS = ("csv", "tb", "wandb", "clearml", "comet") # *.csv, TensorBoard, Weights & Biases, ClearML
|
||||
RANK = int(os.getenv("RANK", -1))
|
||||
|
||||
try:
|
||||
from torch.utils.tensorboard import SummaryWriter
|
||||
except ImportError:
|
||||
|
||||
def SummaryWriter(*args):
|
||||
"""Fall back to SummaryWriter returning None if TensorBoard is not installed."""
|
||||
return None # None = SummaryWriter(str)
|
||||
|
||||
|
||||
try:
|
||||
import wandb
|
||||
|
||||
assert hasattr(wandb, "__version__") # verify package import not local dir
|
||||
if pkg.parse_version(wandb.__version__) >= pkg.parse_version("0.12.2") and RANK in {0, -1}:
|
||||
try:
|
||||
wandb_login_success = wandb.login(timeout=30)
|
||||
except wandb.errors.UsageError: # known non-TTY terminal issue
|
||||
wandb_login_success = False
|
||||
if not wandb_login_success:
|
||||
wandb = None
|
||||
except (ImportError, AssertionError):
|
||||
wandb = None
|
||||
|
||||
try:
|
||||
import clearml
|
||||
|
||||
assert hasattr(clearml, "__version__") # verify package import not local dir
|
||||
except (ImportError, AssertionError):
|
||||
clearml = None
|
||||
|
||||
try:
|
||||
if RANK in {0, -1}:
|
||||
import comet_ml
|
||||
|
||||
assert hasattr(comet_ml, "__version__") # verify package import not local dir
|
||||
from utils.loggers.comet import CometLogger
|
||||
|
||||
else:
|
||||
comet_ml = None
|
||||
except (ImportError, AssertionError):
|
||||
comet_ml = None
|
||||
|
||||
|
||||
def _json_default(value):
|
||||
"""
|
||||
Format `value` for JSON serialization (e.g. unwrap tensors).
|
||||
|
||||
Fall back to strings.
|
||||
"""
|
||||
if isinstance(value, torch.Tensor):
|
||||
try:
|
||||
value = value.item()
|
||||
except ValueError: # "only one element tensors can be converted to Python scalars"
|
||||
pass
|
||||
return value if isinstance(value, float) else str(value)
|
||||
|
||||
|
||||
class Loggers:
|
||||
"""Initializes and manages various logging utilities for tracking YOLOv5 training and validation metrics."""
|
||||
|
||||
def __init__(self, save_dir=None, weights=None, opt=None, hyp=None, logger=None, include=LOGGERS):
|
||||
"""Initializes loggers for YOLOv5 training and validation metrics, paths, and options."""
|
||||
self.save_dir = save_dir
|
||||
self.weights = weights
|
||||
self.opt = opt
|
||||
self.hyp = hyp
|
||||
self.plots = not opt.noplots # plot results
|
||||
self.logger = logger # for printing results to console
|
||||
self.include = include
|
||||
self.keys = [
|
||||
"train/box_loss",
|
||||
"train/obj_loss",
|
||||
"train/cls_loss", # train loss
|
||||
"metrics/precision",
|
||||
"metrics/recall",
|
||||
"metrics/mAP_0.5",
|
||||
"metrics/mAP_0.5:0.95", # metrics
|
||||
"val/box_loss",
|
||||
"val/obj_loss",
|
||||
"val/cls_loss", # val loss
|
||||
"x/lr0",
|
||||
"x/lr1",
|
||||
"x/lr2",
|
||||
] # params
|
||||
self.best_keys = ["best/epoch", "best/precision", "best/recall", "best/mAP_0.5", "best/mAP_0.5:0.95"]
|
||||
for k in LOGGERS:
|
||||
setattr(self, k, None) # init empty logger dictionary
|
||||
self.csv = True # always log to csv
|
||||
self.ndjson_console = "ndjson_console" in self.include # log ndjson to console
|
||||
self.ndjson_file = "ndjson_file" in self.include # log ndjson to file
|
||||
|
||||
# Messages
|
||||
if not comet_ml:
|
||||
prefix = colorstr("Comet: ")
|
||||
s = f"{prefix}run 'pip install comet_ml' to automatically track and visualize YOLOv5 🚀 runs in Comet"
|
||||
self.logger.info(s)
|
||||
# TensorBoard
|
||||
s = self.save_dir
|
||||
if "tb" in self.include and not self.opt.evolve:
|
||||
prefix = colorstr("TensorBoard: ")
|
||||
self.logger.info(f"{prefix}Start with 'tensorboard --logdir {s.parent}', view at http://localhost:6006/")
|
||||
self.tb = SummaryWriter(str(s))
|
||||
|
||||
# W&B
|
||||
if wandb and "wandb" in self.include:
|
||||
self.opt.hyp = self.hyp # add hyperparameters
|
||||
self.wandb = WandbLogger(self.opt)
|
||||
else:
|
||||
self.wandb = None
|
||||
|
||||
# ClearML
|
||||
if clearml and "clearml" in self.include:
|
||||
try:
|
||||
self.clearml = ClearmlLogger(self.opt, self.hyp)
|
||||
except Exception:
|
||||
self.clearml = None
|
||||
prefix = colorstr("ClearML: ")
|
||||
LOGGER.warning(
|
||||
f"{prefix}WARNING ⚠️ ClearML is installed but not configured, skipping ClearML logging."
|
||||
f" See https://docs.ultralytics.com/yolov5/tutorials/clearml_logging_integration#readme"
|
||||
)
|
||||
|
||||
else:
|
||||
self.clearml = None
|
||||
|
||||
# Comet
|
||||
if comet_ml and "comet" in self.include:
|
||||
if isinstance(self.opt.resume, str) and self.opt.resume.startswith("comet://"):
|
||||
run_id = self.opt.resume.split("/")[-1]
|
||||
self.comet_logger = CometLogger(self.opt, self.hyp, run_id=run_id)
|
||||
|
||||
else:
|
||||
self.comet_logger = CometLogger(self.opt, self.hyp)
|
||||
|
||||
else:
|
||||
self.comet_logger = None
|
||||
|
||||
@property
|
||||
def remote_dataset(self):
|
||||
"""Fetches dataset dictionary from remote logging services like ClearML, Weights & Biases, or Comet ML."""
|
||||
data_dict = None
|
||||
if self.clearml:
|
||||
data_dict = self.clearml.data_dict
|
||||
if self.wandb:
|
||||
data_dict = self.wandb.data_dict
|
||||
if self.comet_logger:
|
||||
data_dict = self.comet_logger.data_dict
|
||||
|
||||
return data_dict
|
||||
|
||||
def on_train_start(self):
|
||||
"""Initializes the training process for Comet ML logger if it's configured."""
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_train_start()
|
||||
|
||||
def on_pretrain_routine_start(self):
|
||||
"""Invokes pre-training routine start hook for Comet ML logger if available."""
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_pretrain_routine_start()
|
||||
|
||||
def on_pretrain_routine_end(self, labels, names):
|
||||
"""Callback that runs at the end of pre-training routine, logging label plots if enabled."""
|
||||
if self.plots:
|
||||
plot_labels(labels, names, self.save_dir)
|
||||
paths = self.save_dir.glob("*labels*.jpg") # training labels
|
||||
if self.wandb:
|
||||
self.wandb.log({"Labels": [wandb.Image(str(x), caption=x.name) for x in paths]})
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_pretrain_routine_end(paths)
|
||||
if self.clearml:
|
||||
for path in paths:
|
||||
self.clearml.log_plot(title=path.stem, plot_path=path)
|
||||
|
||||
def on_train_batch_end(self, model, ni, imgs, targets, paths, vals):
|
||||
"""Logs training batch end events, plots images, and updates external loggers with batch-end data."""
|
||||
log_dict = dict(zip(self.keys[:3], vals))
|
||||
# Callback runs on train batch end
|
||||
# ni: number integrated batches (since train start)
|
||||
if self.plots:
|
||||
if ni < 3:
|
||||
f = self.save_dir / f"train_batch{ni}.jpg" # filename
|
||||
plot_images(imgs, targets, paths, f)
|
||||
if ni == 0 and self.tb and not self.opt.sync_bn:
|
||||
log_tensorboard_graph(self.tb, model, imgsz=(self.opt.imgsz, self.opt.imgsz))
|
||||
if ni == 10 and (self.wandb or self.clearml):
|
||||
files = sorted(self.save_dir.glob("train*.jpg"))
|
||||
if self.wandb:
|
||||
self.wandb.log({"Mosaics": [wandb.Image(str(f), caption=f.name) for f in files if f.exists()]})
|
||||
if self.clearml:
|
||||
self.clearml.log_debug_samples(files, title="Mosaics")
|
||||
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_train_batch_end(log_dict, step=ni)
|
||||
|
||||
def on_train_epoch_end(self, epoch):
|
||||
"""Callback that updates the current epoch in Weights & Biases at the end of a training epoch."""
|
||||
if self.wandb:
|
||||
self.wandb.current_epoch = epoch + 1
|
||||
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_train_epoch_end(epoch)
|
||||
|
||||
def on_val_start(self):
|
||||
"""Callback that signals the start of a validation phase to the Comet logger."""
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_val_start()
|
||||
|
||||
def on_val_image_end(self, pred, predn, path, names, im):
|
||||
"""Callback that logs a validation image and its predictions to WandB or ClearML."""
|
||||
if self.wandb:
|
||||
self.wandb.val_one_image(pred, predn, path, names, im)
|
||||
if self.clearml:
|
||||
self.clearml.log_image_with_boxes(path, pred, names, im)
|
||||
|
||||
def on_val_batch_end(self, batch_i, im, targets, paths, shapes, out):
|
||||
"""Logs validation batch results to Comet ML during training at the end of each validation batch."""
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_val_batch_end(batch_i, im, targets, paths, shapes, out)
|
||||
|
||||
def on_val_end(self, nt, tp, fp, p, r, f1, ap, ap50, ap_class, confusion_matrix):
|
||||
"""Logs validation results to WandB or ClearML at the end of the validation process."""
|
||||
if self.wandb or self.clearml:
|
||||
files = sorted(self.save_dir.glob("val*.jpg"))
|
||||
if self.wandb:
|
||||
self.wandb.log({"Validation": [wandb.Image(str(f), caption=f.name) for f in files]})
|
||||
if self.clearml:
|
||||
self.clearml.log_debug_samples(files, title="Validation")
|
||||
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_val_end(nt, tp, fp, p, r, f1, ap, ap50, ap_class, confusion_matrix)
|
||||
|
||||
def on_fit_epoch_end(self, vals, epoch, best_fitness, fi):
|
||||
"""Callback that logs metrics and saves them to CSV or NDJSON at the end of each fit (train+val) epoch."""
|
||||
x = dict(zip(self.keys, vals))
|
||||
if self.csv:
|
||||
file = self.save_dir / "results.csv"
|
||||
n = len(x) + 1 # number of cols
|
||||
s = "" if file.exists() else (("%20s," * n % tuple(["epoch"] + self.keys)).rstrip(",") + "\n") # add header
|
||||
with open(file, "a") as f:
|
||||
f.write(s + ("%20.5g," * n % tuple([epoch] + vals)).rstrip(",") + "\n")
|
||||
if self.ndjson_console or self.ndjson_file:
|
||||
json_data = json.dumps(dict(epoch=epoch, **x), default=_json_default)
|
||||
if self.ndjson_console:
|
||||
print(json_data)
|
||||
if self.ndjson_file:
|
||||
file = self.save_dir / "results.ndjson"
|
||||
with open(file, "a") as f:
|
||||
print(json_data, file=f)
|
||||
|
||||
if self.tb:
|
||||
for k, v in x.items():
|
||||
self.tb.add_scalar(k, v, epoch)
|
||||
elif self.clearml: # log to ClearML if TensorBoard not used
|
||||
self.clearml.log_scalars(x, epoch)
|
||||
|
||||
if self.wandb:
|
||||
if best_fitness == fi:
|
||||
best_results = [epoch] + vals[3:7]
|
||||
for i, name in enumerate(self.best_keys):
|
||||
self.wandb.wandb_run.summary[name] = best_results[i] # log best results in the summary
|
||||
self.wandb.log(x)
|
||||
self.wandb.end_epoch()
|
||||
|
||||
if self.clearml:
|
||||
self.clearml.current_epoch_logged_images = set() # reset epoch image limit
|
||||
self.clearml.current_epoch += 1
|
||||
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_fit_epoch_end(x, epoch=epoch)
|
||||
|
||||
def on_model_save(self, last, epoch, final_epoch, best_fitness, fi):
|
||||
"""Callback that handles model saving events, logging to Weights & Biases or ClearML if enabled."""
|
||||
if (epoch + 1) % self.opt.save_period == 0 and not final_epoch and self.opt.save_period != -1:
|
||||
if self.wandb:
|
||||
self.wandb.log_model(last.parent, self.opt, epoch, fi, best_model=best_fitness == fi)
|
||||
if self.clearml:
|
||||
self.clearml.task.update_output_model(
|
||||
model_path=str(last), model_name="Latest Model", auto_delete_file=False
|
||||
)
|
||||
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_model_save(last, epoch, final_epoch, best_fitness, fi)
|
||||
|
||||
def on_train_end(self, last, best, epoch, results):
|
||||
"""Callback that runs at the end of training to save plots and log results."""
|
||||
if self.plots:
|
||||
plot_results(file=self.save_dir / "results.csv") # save results.png
|
||||
files = ["results.png", "confusion_matrix.png", *(f"{x}_curve.png" for x in ("F1", "PR", "P", "R"))]
|
||||
files = [(self.save_dir / f) for f in files if (self.save_dir / f).exists()] # filter
|
||||
self.logger.info(f"Results saved to {colorstr('bold', self.save_dir)}")
|
||||
|
||||
if self.tb and not self.clearml: # These images are already captured by ClearML by now, we don't want doubles
|
||||
for f in files:
|
||||
self.tb.add_image(f.stem, cv2.imread(str(f))[..., ::-1], epoch, dataformats="HWC")
|
||||
|
||||
if self.wandb:
|
||||
self.wandb.log(dict(zip(self.keys[3:10], results)))
|
||||
self.wandb.log({"Results": [wandb.Image(str(f), caption=f.name) for f in files]})
|
||||
# Calling wandb.log. TODO: Refactor this into WandbLogger.log_model
|
||||
if not self.opt.evolve:
|
||||
wandb.log_artifact(
|
||||
str(best if best.exists() else last),
|
||||
type="model",
|
||||
name=f"run_{self.wandb.wandb_run.id}_model",
|
||||
aliases=["latest", "best", "stripped"],
|
||||
)
|
||||
self.wandb.finish_run()
|
||||
|
||||
if self.clearml and not self.opt.evolve:
|
||||
self.clearml.log_summary(dict(zip(self.keys[3:10], results)))
|
||||
[self.clearml.log_plot(title=f.stem, plot_path=f) for f in files]
|
||||
self.clearml.log_model(
|
||||
str(best if best.exists() else last), "Best Model" if best.exists() else "Last Model", epoch
|
||||
)
|
||||
|
||||
if self.comet_logger:
|
||||
final_results = dict(zip(self.keys[3:10], results))
|
||||
self.comet_logger.on_train_end(files, self.save_dir, last, best, epoch, final_results)
|
||||
|
||||
def on_params_update(self, params: dict):
|
||||
"""Updates experiment hyperparameters or configurations in WandB, Comet, or ClearML."""
|
||||
if self.wandb:
|
||||
self.wandb.wandb_run.config.update(params, allow_val_change=True)
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_params_update(params)
|
||||
if self.clearml:
|
||||
self.clearml.task.connect(params)
|
||||
|
||||
|
||||
class GenericLogger:
|
||||
"""
|
||||
YOLOv5 General purpose logger for non-task specific logging
|
||||
Usage: from utils.loggers import GenericLogger; logger = GenericLogger(...).
|
||||
|
||||
Arguments:
|
||||
opt: Run arguments
|
||||
console_logger: Console logger
|
||||
include: loggers to include
|
||||
"""
|
||||
|
||||
def __init__(self, opt, console_logger, include=("tb", "wandb", "clearml")):
|
||||
"""Initializes a generic logger with optional TensorBoard, W&B, and ClearML support."""
|
||||
self.save_dir = Path(opt.save_dir)
|
||||
self.include = include
|
||||
self.console_logger = console_logger
|
||||
self.csv = self.save_dir / "results.csv" # CSV logger
|
||||
if "tb" in self.include:
|
||||
prefix = colorstr("TensorBoard: ")
|
||||
self.console_logger.info(
|
||||
f"{prefix}Start with 'tensorboard --logdir {self.save_dir.parent}', view at http://localhost:6006/"
|
||||
)
|
||||
self.tb = SummaryWriter(str(self.save_dir))
|
||||
|
||||
if wandb and "wandb" in self.include:
|
||||
self.wandb = wandb.init(
|
||||
project=web_project_name(str(opt.project)), name=None if opt.name == "exp" else opt.name, config=opt
|
||||
)
|
||||
else:
|
||||
self.wandb = None
|
||||
|
||||
if clearml and "clearml" in self.include:
|
||||
try:
|
||||
# Hyp is not available in classification mode
|
||||
hyp = {} if "hyp" not in opt else opt.hyp
|
||||
self.clearml = ClearmlLogger(opt, hyp)
|
||||
except Exception:
|
||||
self.clearml = None
|
||||
prefix = colorstr("ClearML: ")
|
||||
LOGGER.warning(
|
||||
f"{prefix}WARNING ⚠️ ClearML is installed but not configured, skipping ClearML logging."
|
||||
f" See https://docs.ultralytics.com/yolov5/tutorials/clearml_logging_integration"
|
||||
)
|
||||
else:
|
||||
self.clearml = None
|
||||
|
||||
def log_metrics(self, metrics, epoch):
|
||||
"""Logs metrics to CSV, TensorBoard, W&B, and ClearML; `metrics` is a dict, `epoch` is an int."""
|
||||
if self.csv:
|
||||
keys, vals = list(metrics.keys()), list(metrics.values())
|
||||
n = len(metrics) + 1 # number of cols
|
||||
s = "" if self.csv.exists() else (("%23s," * n % tuple(["epoch"] + keys)).rstrip(",") + "\n") # header
|
||||
with open(self.csv, "a") as f:
|
||||
f.write(s + ("%23.5g," * n % tuple([epoch] + vals)).rstrip(",") + "\n")
|
||||
|
||||
if self.tb:
|
||||
for k, v in metrics.items():
|
||||
self.tb.add_scalar(k, v, epoch)
|
||||
|
||||
if self.wandb:
|
||||
self.wandb.log(metrics, step=epoch)
|
||||
|
||||
if self.clearml:
|
||||
self.clearml.log_scalars(metrics, epoch)
|
||||
|
||||
def log_images(self, files, name="Images", epoch=0):
|
||||
"""Logs images to all loggers with optional naming and epoch specification."""
|
||||
files = [Path(f) for f in (files if isinstance(files, (tuple, list)) else [files])] # to Path
|
||||
files = [f for f in files if f.exists()] # filter by exists
|
||||
|
||||
if self.tb:
|
||||
for f in files:
|
||||
self.tb.add_image(f.stem, cv2.imread(str(f))[..., ::-1], epoch, dataformats="HWC")
|
||||
|
||||
if self.wandb:
|
||||
self.wandb.log({name: [wandb.Image(str(f), caption=f.name) for f in files]}, step=epoch)
|
||||
|
||||
if self.clearml:
|
||||
if name == "Results":
|
||||
[self.clearml.log_plot(f.stem, f) for f in files]
|
||||
else:
|
||||
self.clearml.log_debug_samples(files, title=name)
|
||||
|
||||
def log_graph(self, model, imgsz=(640, 640)):
|
||||
"""Logs model graph to all configured loggers with specified input image size."""
|
||||
if self.tb:
|
||||
log_tensorboard_graph(self.tb, model, imgsz)
|
||||
|
||||
def log_model(self, model_path, epoch=0, metadata=None):
|
||||
"""Logs the model to all configured loggers with optional epoch and metadata."""
|
||||
if metadata is None:
|
||||
metadata = {}
|
||||
# Log model to all loggers
|
||||
if self.wandb:
|
||||
art = wandb.Artifact(name=f"run_{wandb.run.id}_model", type="model", metadata=metadata)
|
||||
art.add_file(str(model_path))
|
||||
wandb.log_artifact(art)
|
||||
if self.clearml:
|
||||
self.clearml.log_model(model_path=model_path, model_name=model_path.stem)
|
||||
|
||||
def update_params(self, params):
|
||||
"""Updates logged parameters in WandB and/or ClearML if enabled."""
|
||||
if self.wandb:
|
||||
wandb.run.config.update(params, allow_val_change=True)
|
||||
if self.clearml:
|
||||
self.clearml.task.connect(params)
|
||||
|
||||
|
||||
def log_tensorboard_graph(tb, model, imgsz=(640, 640)):
|
||||
"""Logs the model graph to TensorBoard with specified image size and model."""
|
||||
try:
|
||||
p = next(model.parameters()) # for device, type
|
||||
imgsz = (imgsz, imgsz) if isinstance(imgsz, int) else imgsz # expand
|
||||
im = torch.zeros((1, 3, *imgsz)).to(p.device).type_as(p) # input image (WARNING: must be zeros, not empty)
|
||||
with warnings.catch_warnings():
|
||||
warnings.simplefilter("ignore") # suppress jit trace warning
|
||||
tb.add_graph(torch.jit.trace(de_parallel(model), im, strict=False), [])
|
||||
except Exception as e:
|
||||
LOGGER.warning(f"WARNING ⚠️ TensorBoard graph visualization failure {e}")
|
||||
|
||||
|
||||
def web_project_name(project):
|
||||
"""Converts a local project name to a standardized web project name with optional suffixes."""
|
||||
if not project.startswith("runs/train"):
|
||||
return project
|
||||
suffix = "-Classify" if project.endswith("-cls") else "-Segment" if project.endswith("-seg") else ""
|
||||
return f"YOLOv5{suffix}"
|
||||
@@ -1,222 +0,0 @@
|
||||
# ClearML Integration
|
||||
|
||||
<img align="center" src="https://github.com/thepycoder/clearml_screenshots/raw/main/logos_dark.png#gh-light-mode-only" alt="Clear|ML"><img align="center" src="https://github.com/thepycoder/clearml_screenshots/raw/main/logos_light.png#gh-dark-mode-only" alt="Clear|ML">
|
||||
|
||||
## About ClearML
|
||||
|
||||
[ClearML](https://clear.ml/) is an [open-source](https://github.com/clearml/clearml) toolbox designed to save you time ⏱️.
|
||||
|
||||
🔨 Track every YOLOv5 training run in the <b>experiment manager</b>
|
||||
|
||||
🔧 Version and easily access your custom training data with the integrated ClearML <b>Data Versioning Tool</b>
|
||||
|
||||
🔦 <b>Remotely train and monitor</b> your YOLOv5 training runs using ClearML Agent
|
||||
|
||||
🔬 Get the very best mAP using ClearML <b>Hyperparameter Optimization</b>
|
||||
|
||||
🔭 Turn your newly trained <b>YOLOv5 model into an API</b> with just a few commands using ClearML Serving
|
||||
|
||||
And so much more. It's up to you how many of these tools you want to use, you can stick to the experiment manager, or chain them all together into an impressive pipeline!
|
||||
|
||||
|
||||
|
||||
## 🦾 Setting Things Up
|
||||
|
||||
To keep track of your experiments and/or data, ClearML needs to communicate to a server. You have 2 options to get one:
|
||||
|
||||
Either sign up for free to the [ClearML Hosted Service](https://clear.ml/) or you can set up your own server, see [here](https://clear.ml/docs/latest/docs/deploying_clearml/clearml_server). Even the server is open-source, so even if you're dealing with sensitive data, you should be good to go!
|
||||
|
||||
1. Install the `clearml` python package:
|
||||
|
||||
```bash
|
||||
pip install clearml
|
||||
```
|
||||
|
||||
2. Connect the ClearML SDK to the server by [creating credentials](https://app.clear.ml/settings/workspace-configuration) (go right top to Settings -> Workspace -> Create new credentials), then execute the command below and follow the instructions:
|
||||
|
||||
```bash
|
||||
clearml-init
|
||||
```
|
||||
|
||||
That's it! You're done 😎
|
||||
|
||||
## 🚀 Training YOLOv5 With ClearML
|
||||
|
||||
To enable ClearML experiment tracking, simply install the ClearML pip package.
|
||||
|
||||
```bash
|
||||
pip install clearml>=1.2.0
|
||||
```
|
||||
|
||||
This will enable integration with the YOLOv5 training script. Every training run from now on, will be captured and stored by the ClearML experiment manager.
|
||||
|
||||
If you want to change the `project_name` or `task_name`, use the `--project` and `--name` arguments of the `train.py` script, by default the project will be called `YOLOv5` and the task `Training`. PLEASE NOTE: ClearML uses `/` as a delimiter for subprojects, so be careful when using `/` in your project name!
|
||||
|
||||
```bash
|
||||
python train.py --img 640 --batch 16 --epochs 3 --data coco128.yaml --weights yolov5s.pt --cache
|
||||
```
|
||||
|
||||
or with custom project and task name:
|
||||
|
||||
```bash
|
||||
python train.py --project my_project --name my_training --img 640 --batch 16 --epochs 3 --data coco128.yaml --weights yolov5s.pt --cache
|
||||
```
|
||||
|
||||
This will capture:
|
||||
|
||||
- Source code + uncommitted changes
|
||||
- Installed packages
|
||||
- (Hyper)parameters
|
||||
- Model files (use `--save-period n` to save a checkpoint every n epochs)
|
||||
- Console output
|
||||
- Scalars (mAP_0.5, mAP_0.5:0.95, precision, recall, losses, learning rates, ...)
|
||||
- General info such as machine details, runtime, creation date etc.
|
||||
- All produced plots such as label correlogram and confusion matrix
|
||||
- Images with bounding boxes per epoch
|
||||
- Mosaic per epoch
|
||||
- Validation images per epoch
|
||||
- ...
|
||||
|
||||
That's a lot right? 🤯 Now, we can visualize all of this information in the ClearML UI to get an overview of our training progress. Add custom columns to the table view (such as e.g. mAP_0.5) so you can easily sort on the best performing model. Or select multiple experiments and directly compare them!
|
||||
|
||||
There even more we can do with all of this information, like hyperparameter optimization and remote execution, so keep reading if you want to see how that works!
|
||||
|
||||
## 🔗 Dataset Version Management
|
||||
|
||||
Versioning your data separately from your code is generally a good idea and makes it easy to acquire the latest version too. This repository supports supplying a dataset version ID, and it will make sure to get the data if it's not there yet. Next to that, this workflow also saves the used dataset ID as part of the task parameters, so you will always know for sure which data was used in which experiment!
|
||||
|
||||
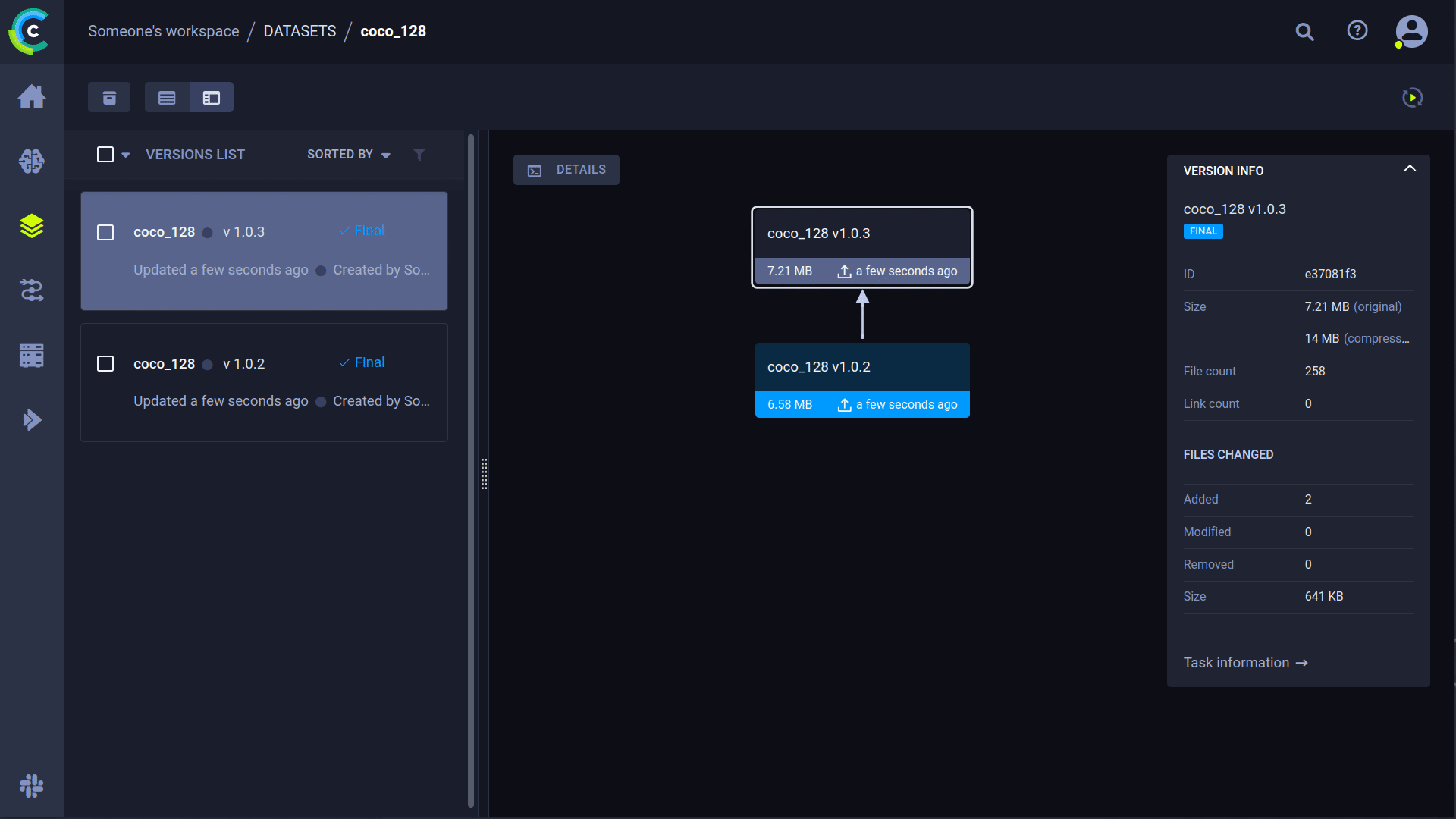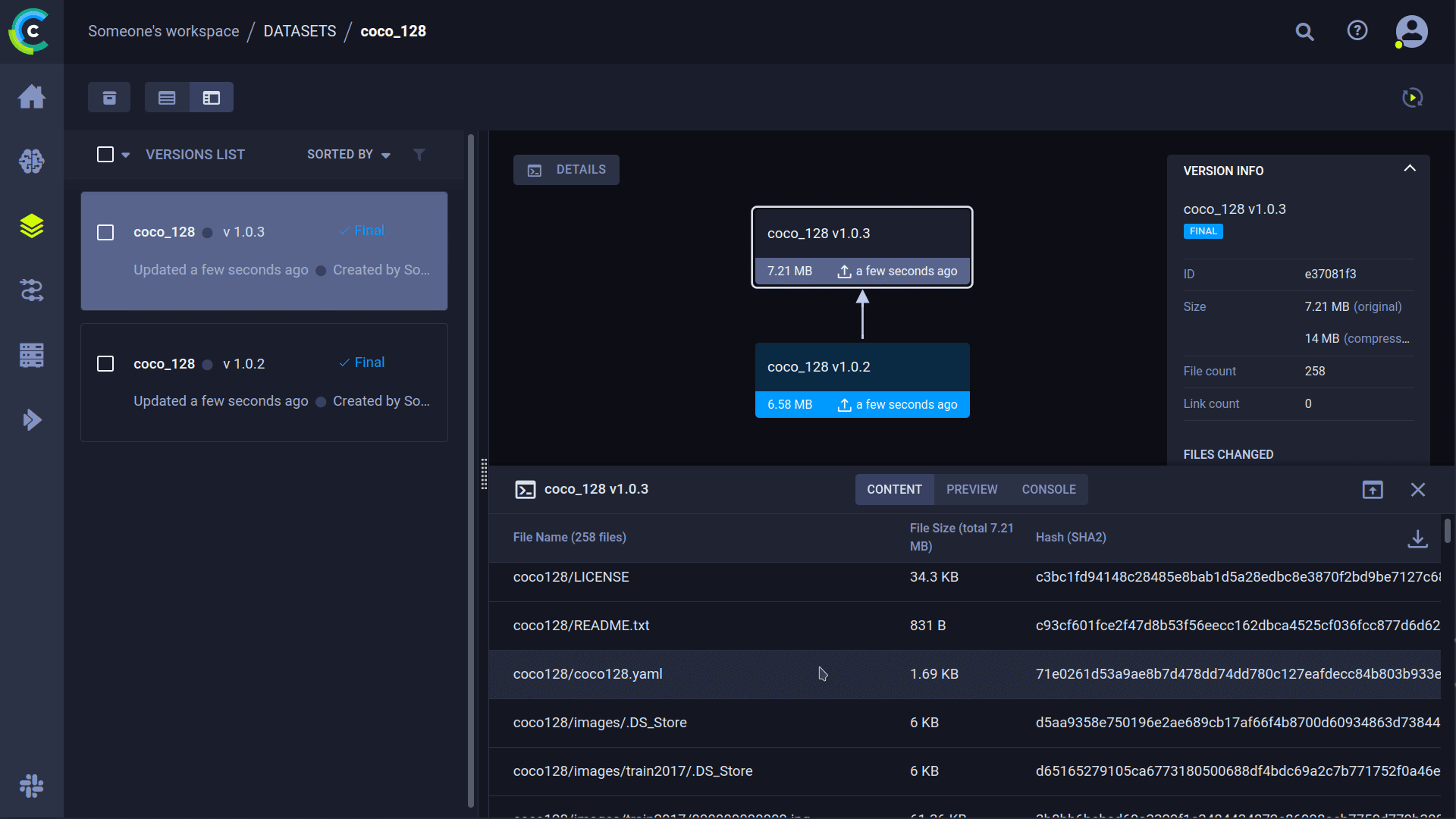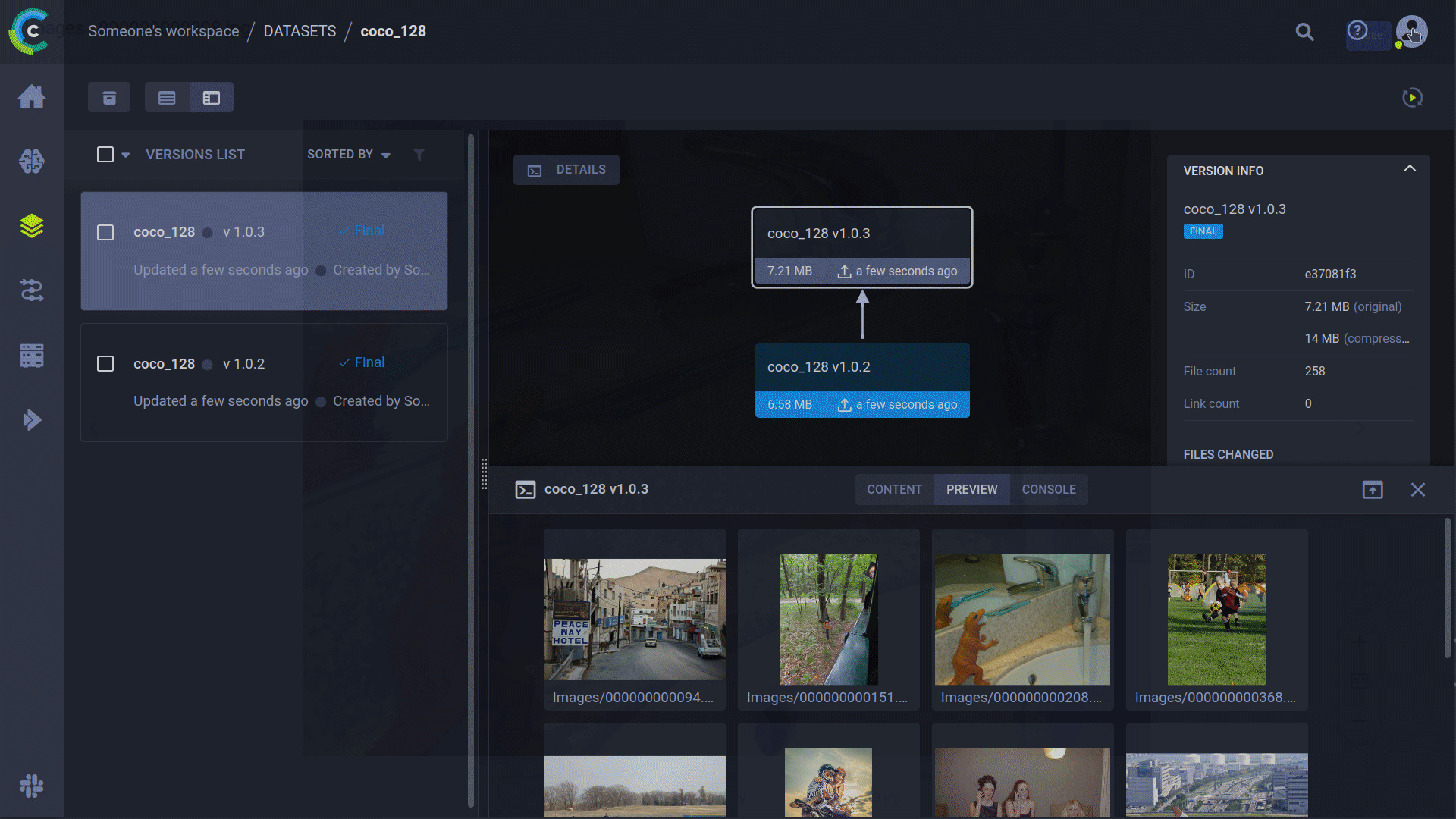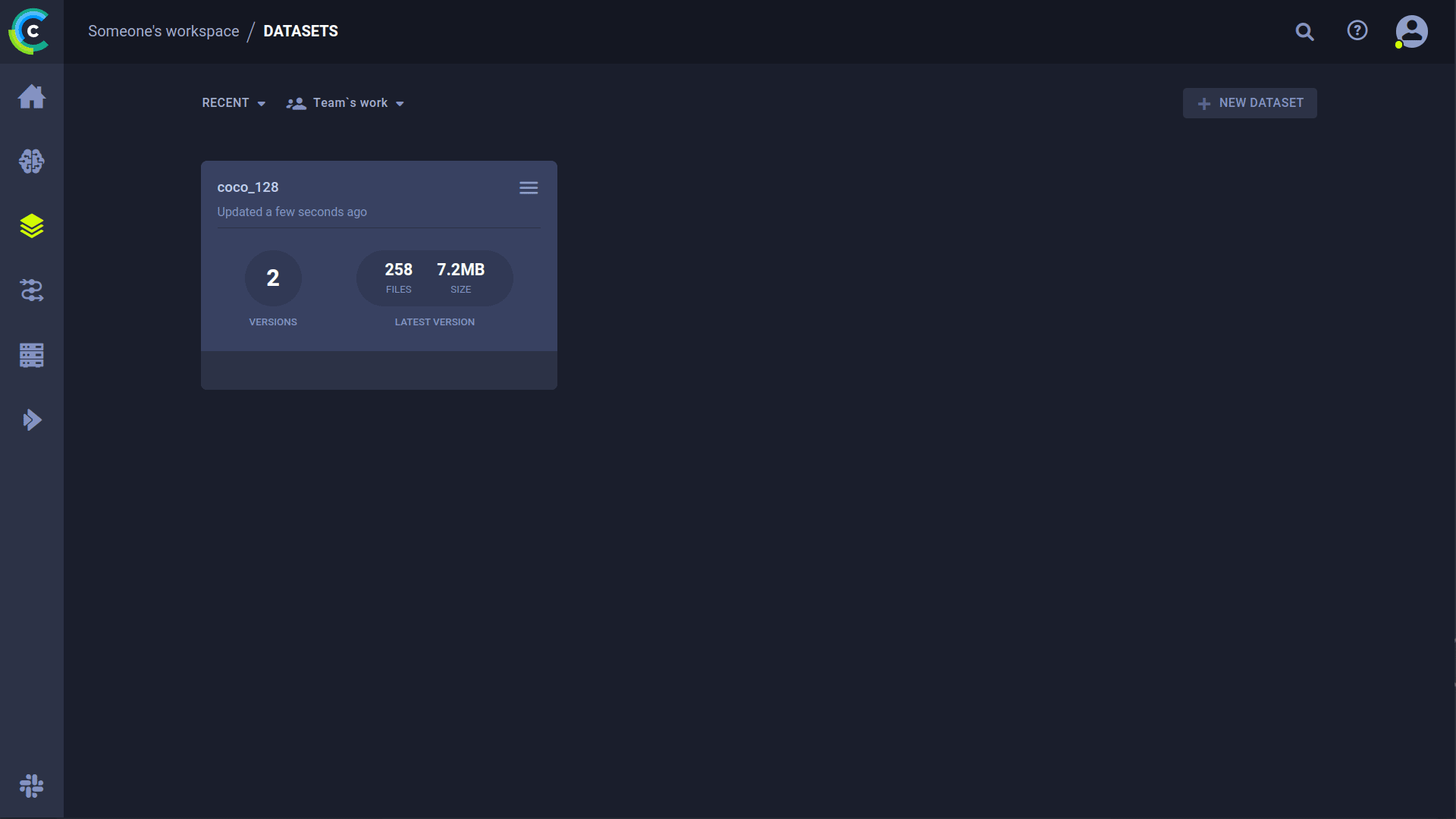
|
||||
|
||||
### Prepare Your Dataset
|
||||
|
||||
The YOLOv5 repository supports a number of different datasets by using yaml files containing their information. By default datasets are downloaded to the `../datasets` folder in relation to the repository root folder. So if you downloaded the `coco128` dataset using the link in the yaml or with the scripts provided by yolov5, you get this folder structure:
|
||||
|
||||
```
|
||||
..
|
||||
|_ yolov5
|
||||
|_ datasets
|
||||
|_ coco128
|
||||
|_ images
|
||||
|_ labels
|
||||
|_ LICENSE
|
||||
|_ README.txt
|
||||
```
|
||||
|
||||
But this can be any dataset you wish. Feel free to use your own, as long as you keep to this folder structure.
|
||||
|
||||
Next, ⚠️**copy the corresponding yaml file to the root of the dataset folder**⚠️. This yaml files contains the information ClearML will need to properly use the dataset. You can make this yourself too, of course, just follow the structure of the example yamls.
|
||||
|
||||
Basically we need the following keys: `path`, `train`, `test`, `val`, `nc`, `names`.
|
||||
|
||||
```
|
||||
..
|
||||
|_ yolov5
|
||||
|_ datasets
|
||||
|_ coco128
|
||||
|_ images
|
||||
|_ labels
|
||||
|_ coco128.yaml # <---- HERE!
|
||||
|_ LICENSE
|
||||
|_ README.txt
|
||||
```
|
||||
|
||||
### Upload Your Dataset
|
||||
|
||||
To get this dataset into ClearML as a versioned dataset, go to the dataset root folder and run the following command:
|
||||
|
||||
```bash
|
||||
cd coco128
|
||||
clearml-data sync --project YOLOv5 --name coco128 --folder .
|
||||
```
|
||||
|
||||
The command `clearml-data sync` is actually a shorthand command. You could also run these commands one after the other:
|
||||
|
||||
```bash
|
||||
# Optionally add --parent <parent_dataset_id> if you want to base
|
||||
# this version on another dataset version, so no duplicate files are uploaded!
|
||||
clearml-data create --name coco128 --project YOLOv5
|
||||
clearml-data add --files .
|
||||
clearml-data close
|
||||
```
|
||||
|
||||
### Run Training Using A ClearML Dataset
|
||||
|
||||
Now that you have a ClearML dataset, you can very simply use it to train custom YOLOv5 🚀 models!
|
||||
|
||||
```bash
|
||||
python train.py --img 640 --batch 16 --epochs 3 --data clearml://<your_dataset_id> --weights yolov5s.pt --cache
|
||||
```
|
||||
|
||||
## 👀 Hyperparameter Optimization
|
||||
|
||||
Now that we have our experiments and data versioned, it's time to take a look at what we can build on top!
|
||||
|
||||
Using the code information, installed packages and environment details, the experiment itself is now **completely reproducible**. In fact, ClearML allows you to clone an experiment and even change its parameters. We can then just rerun it with these new parameters automatically, this is basically what HPO does!
|
||||
|
||||
To **run hyperparameter optimization locally**, we've included a pre-made script for you. Just make sure a training task has been run at least once, so it is in the ClearML experiment manager, we will essentially clone it and change its hyperparameters.
|
||||
|
||||
You'll need to fill in the ID of this `template task` in the script found at `utils/loggers/clearml/hpo.py` and then just run it :) You can change `task.execute_locally()` to `task.execute()` to put it in a ClearML queue and have a remote agent work on it instead.
|
||||
|
||||
```bash
|
||||
# To use optuna, install it first, otherwise you can change the optimizer to just be RandomSearch
|
||||
pip install optuna
|
||||
python utils/loggers/clearml/hpo.py
|
||||
```
|
||||
|
||||
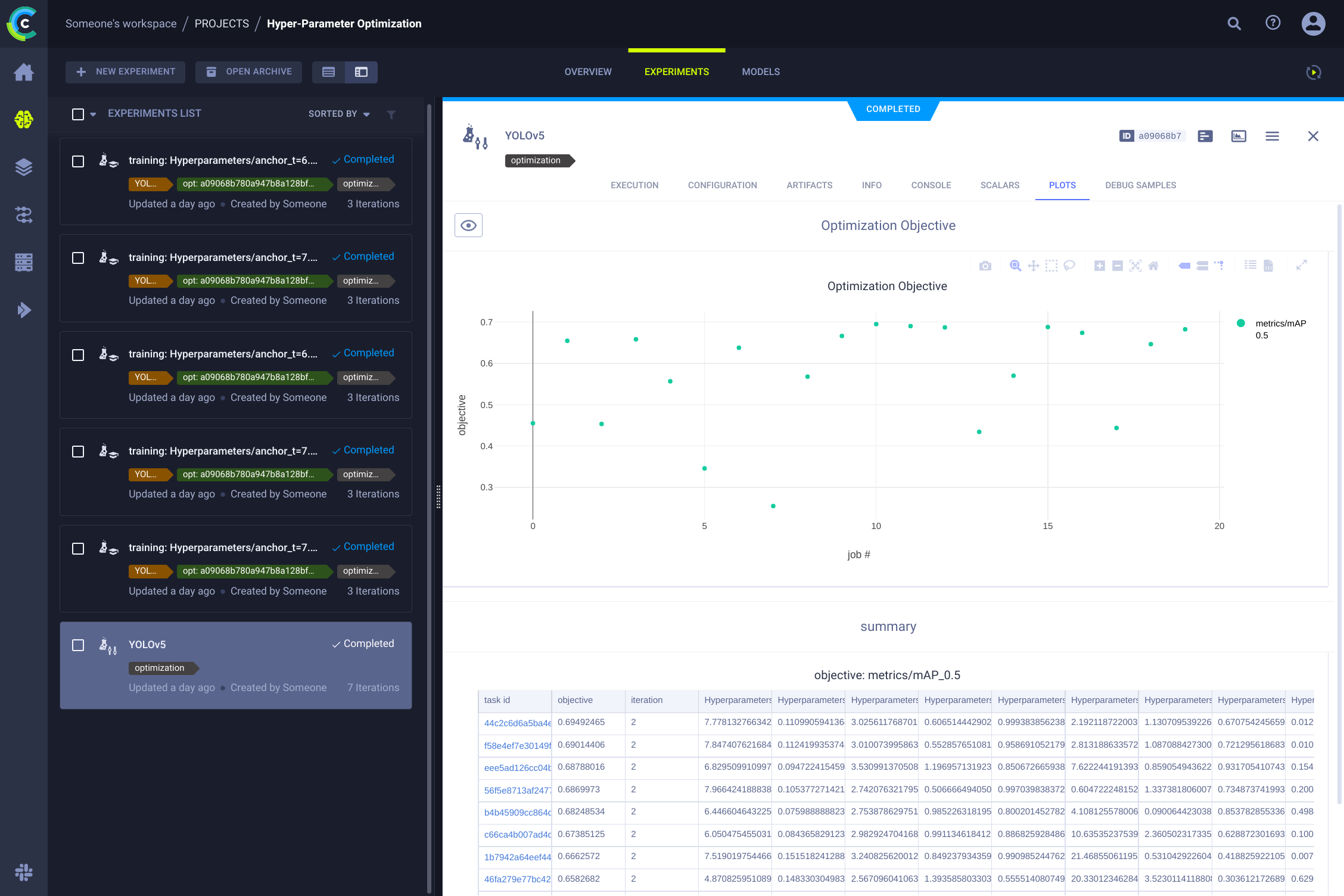
|
||||
|
||||
## 🤯 Remote Execution (advanced)
|
||||
|
||||
Running HPO locally is really handy, but what if we want to run our experiments on a remote machine instead? Maybe you have access to a very powerful GPU machine on-site, or you have some budget to use cloud GPUs. This is where the ClearML Agent comes into play. Check out what the agent can do here:
|
||||
|
||||
- [YouTube video](https://www.youtube.com/watch?v=MX3BrXnaULs&feature=youtu.be)
|
||||
- [Documentation](https://clear.ml/docs/latest/docs/clearml_agent)
|
||||
|
||||
In short: every experiment tracked by the experiment manager contains enough information to reproduce it on a different machine (installed packages, uncommitted changes etc.). So a ClearML agent does just that: it listens to a queue for incoming tasks and when it finds one, it recreates the environment and runs it while still reporting scalars, plots etc. to the experiment manager.
|
||||
|
||||
You can turn any machine (a cloud VM, a local GPU machine, your own laptop ... ) into a ClearML agent by simply running:
|
||||
|
||||
```bash
|
||||
clearml-agent daemon --queue <queues_to_listen_to> [--docker]
|
||||
```
|
||||
|
||||
### Cloning, Editing And Enqueuing
|
||||
|
||||
With our agent running, we can give it some work. Remember from the HPO section that we can clone a task and edit the hyperparameters? We can do that from the interface too!
|
||||
|
||||
🪄 Clone the experiment by right-clicking it
|
||||
|
||||
🎯 Edit the hyperparameters to what you wish them to be
|
||||
|
||||
⏳ Enqueue the task to any of the queues by right-clicking it
|
||||
|
||||
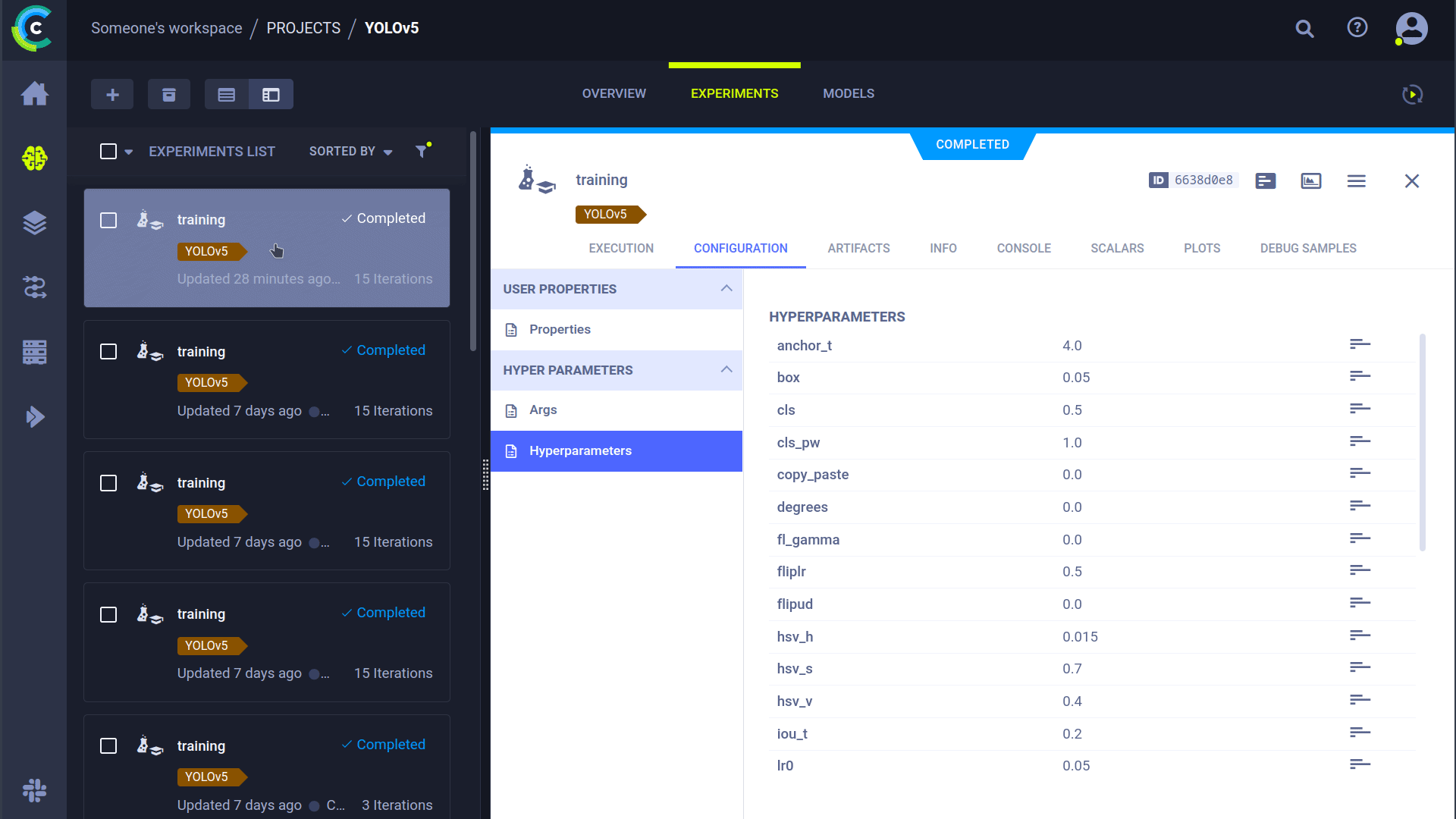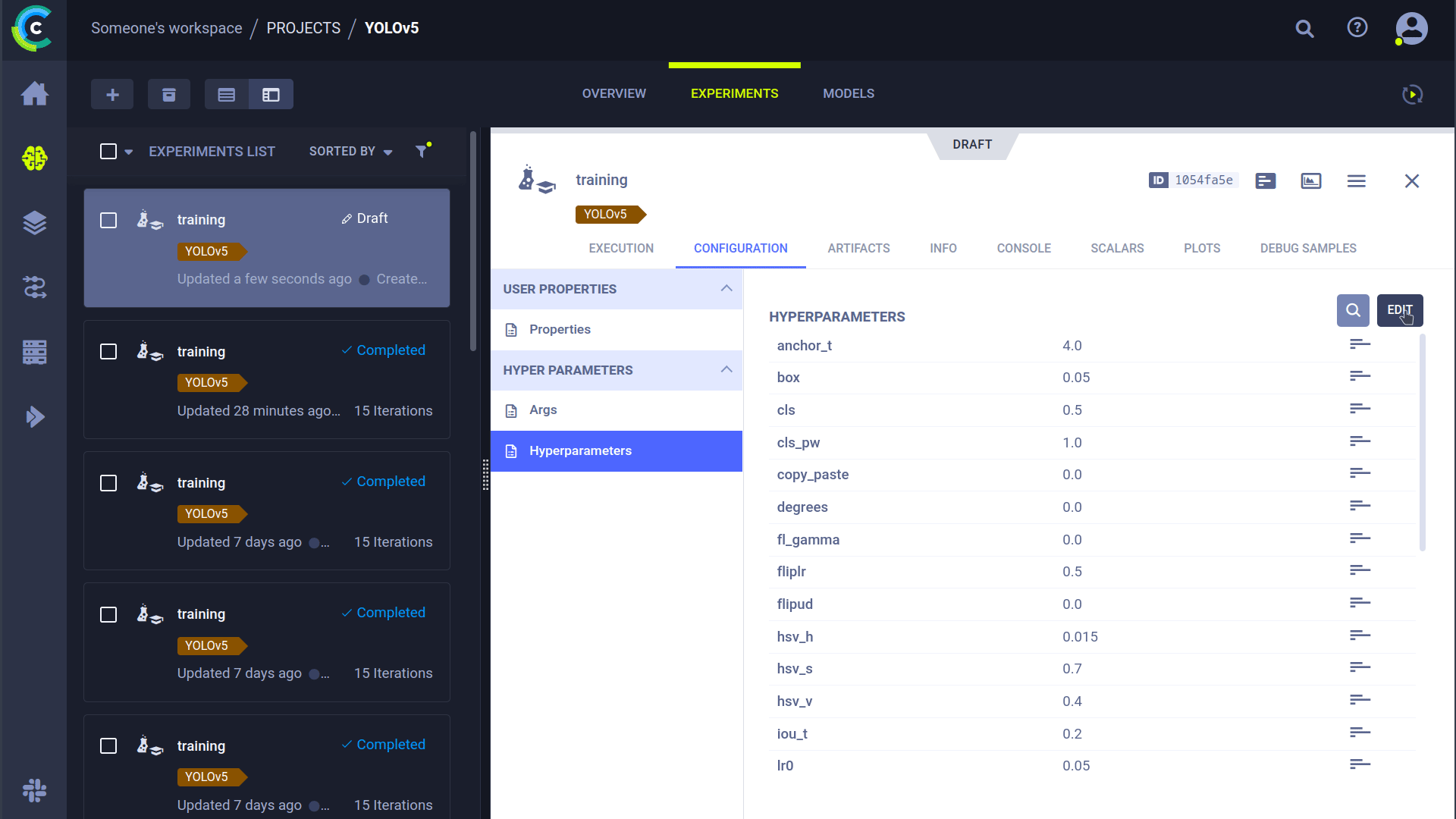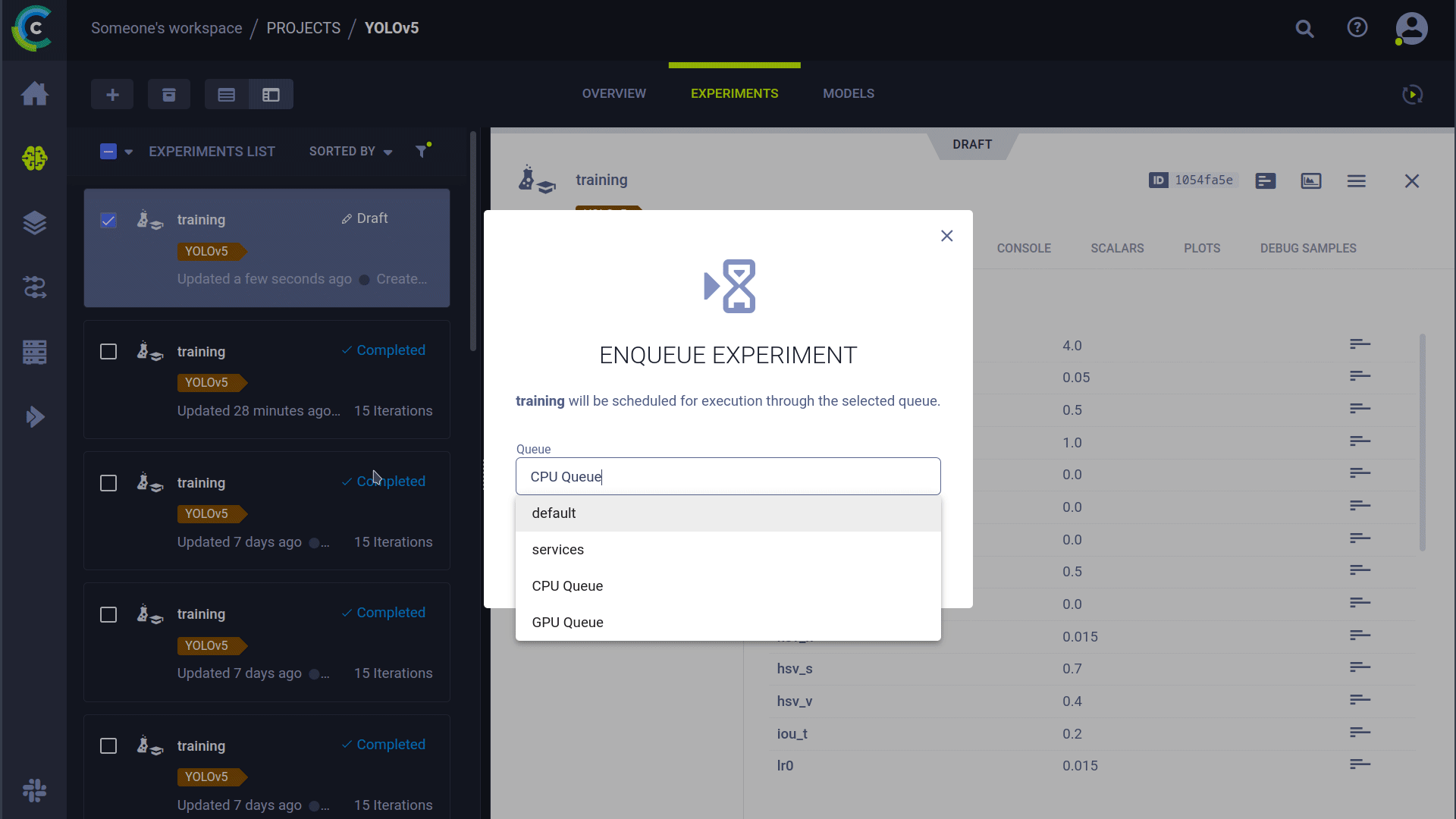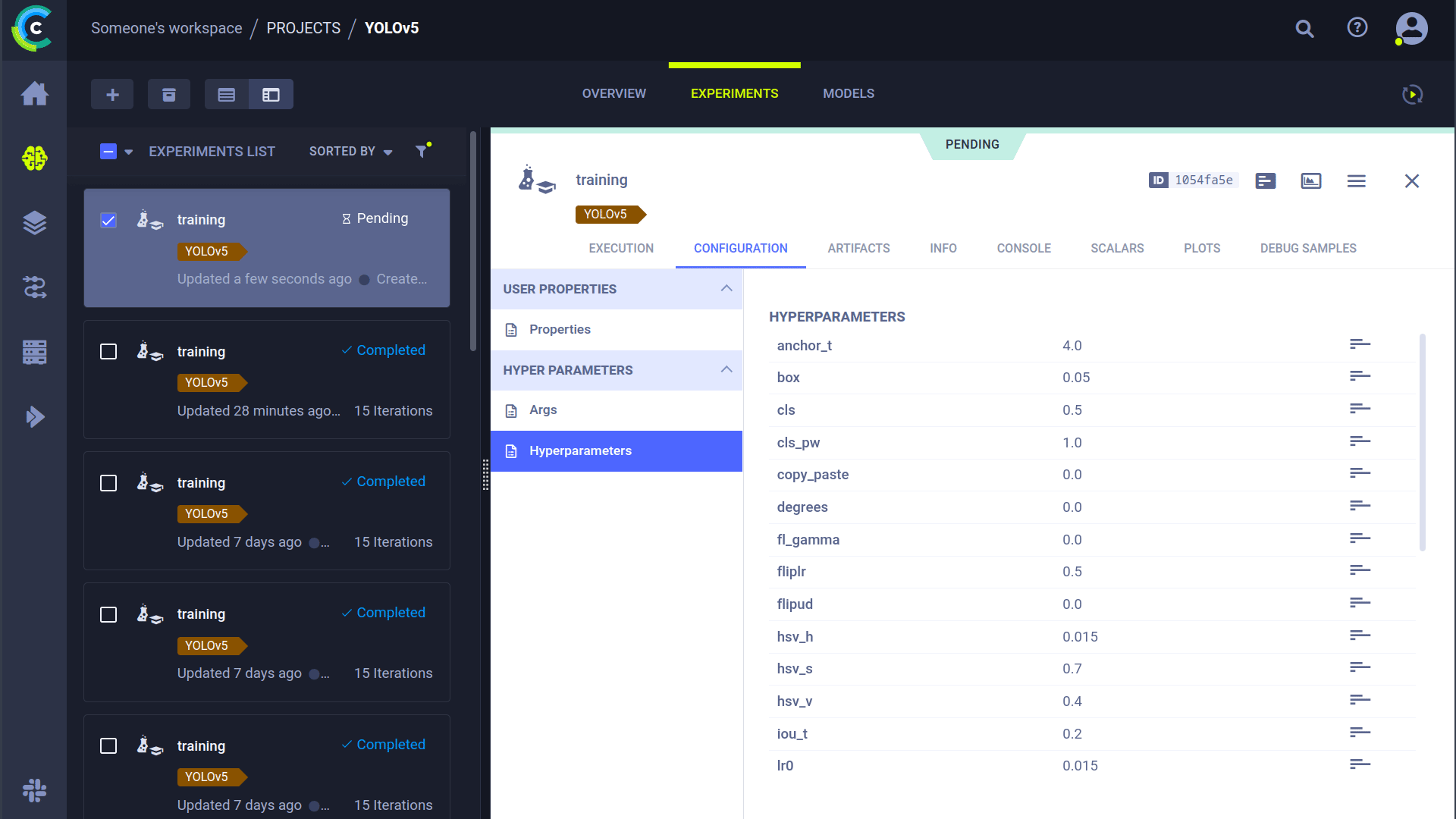
|
||||
|
||||
### Executing A Task Remotely
|
||||
|
||||
Now you can clone a task like we explained above, or simply mark your current script by adding `task.execute_remotely()` and on execution it will be put into a queue, for the agent to start working on!
|
||||
|
||||
To run the YOLOv5 training script remotely, all you have to do is add this line to the training.py script after the clearml logger has been instantiated:
|
||||
|
||||
```python
|
||||
# ...
|
||||
# Loggers
|
||||
data_dict = None
|
||||
if RANK in {-1, 0}:
|
||||
loggers = Loggers(save_dir, weights, opt, hyp, LOGGER) # loggers instance
|
||||
if loggers.clearml:
|
||||
loggers.clearml.task.execute_remotely(queue="my_queue") # <------ ADD THIS LINE
|
||||
# Data_dict is either None is user did not choose for ClearML dataset or is filled in by ClearML
|
||||
data_dict = loggers.clearml.data_dict
|
||||
# ...
|
||||
```
|
||||
|
||||
When running the training script after this change, python will run the script up until that line, after which it will package the code and send it to the queue instead!
|
||||
|
||||
### Autoscaling workers
|
||||
|
||||
ClearML comes with autoscalers too! This tool will automatically spin up new remote machines in the cloud of your choice (AWS, GCP, Azure) and turn them into ClearML agents for you whenever there are experiments detected in the queue. Once the tasks are processed, the autoscaler will automatically shut down the remote machines, and you stop paying!
|
||||
|
||||
Check out the autoscalers getting started video below.
|
||||
|
||||
[](https://youtu.be/j4XVMAaUt3E)
|
||||
@@ -1 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
@@ -1,228 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Main Logger class for ClearML experiment tracking."""
|
||||
|
||||
import glob
|
||||
import re
|
||||
from pathlib import Path
|
||||
|
||||
import matplotlib.image as mpimg
|
||||
import matplotlib.pyplot as plt
|
||||
import numpy as np
|
||||
import yaml
|
||||
from ultralytics.utils.plotting import Annotator, colors
|
||||
|
||||
try:
|
||||
import clearml
|
||||
from clearml import Dataset, Task
|
||||
|
||||
assert hasattr(clearml, "__version__") # verify package import not local dir
|
||||
except (ImportError, AssertionError):
|
||||
clearml = None
|
||||
|
||||
|
||||
def construct_dataset(clearml_info_string):
|
||||
"""Load in a clearml dataset and fill the internal data_dict with its contents."""
|
||||
dataset_id = clearml_info_string.replace("clearml://", "")
|
||||
dataset = Dataset.get(dataset_id=dataset_id)
|
||||
dataset_root_path = Path(dataset.get_local_copy())
|
||||
|
||||
# We'll search for the yaml file definition in the dataset
|
||||
yaml_filenames = list(glob.glob(str(dataset_root_path / "*.yaml")) + glob.glob(str(dataset_root_path / "*.yml")))
|
||||
if len(yaml_filenames) > 1:
|
||||
raise ValueError(
|
||||
"More than one yaml file was found in the dataset root, cannot determine which one contains "
|
||||
"the dataset definition this way."
|
||||
)
|
||||
elif not yaml_filenames:
|
||||
raise ValueError(
|
||||
"No yaml definition found in dataset root path, check that there is a correct yaml file "
|
||||
"inside the dataset root path."
|
||||
)
|
||||
with open(yaml_filenames[0]) as f:
|
||||
dataset_definition = yaml.safe_load(f)
|
||||
|
||||
assert set(dataset_definition.keys()).issuperset({"train", "test", "val", "nc", "names"}), (
|
||||
"The right keys were not found in the yaml file, make sure it at least has the following keys: ('train', 'test', 'val', 'nc', 'names')"
|
||||
)
|
||||
|
||||
data_dict = {
|
||||
"train": (
|
||||
str((dataset_root_path / dataset_definition["train"]).resolve()) if dataset_definition["train"] else None
|
||||
)
|
||||
}
|
||||
data_dict["test"] = (
|
||||
str((dataset_root_path / dataset_definition["test"]).resolve()) if dataset_definition["test"] else None
|
||||
)
|
||||
data_dict["val"] = (
|
||||
str((dataset_root_path / dataset_definition["val"]).resolve()) if dataset_definition["val"] else None
|
||||
)
|
||||
data_dict["nc"] = dataset_definition["nc"]
|
||||
data_dict["names"] = dataset_definition["names"]
|
||||
|
||||
return data_dict
|
||||
|
||||
|
||||
class ClearmlLogger:
|
||||
"""
|
||||
Log training runs, datasets, models, and predictions to ClearML.
|
||||
|
||||
This logger sends information to ClearML at app.clear.ml or to your own hosted server. By default, this information
|
||||
includes hyperparameters, system configuration and metrics, model metrics, code information and basic data metrics
|
||||
and analyses.
|
||||
|
||||
By providing additional command line arguments to train.py, datasets, models and predictions can also be logged.
|
||||
"""
|
||||
|
||||
def __init__(self, opt, hyp):
|
||||
"""
|
||||
- Initialize ClearML Task, this object will capture the experiment
|
||||
- Upload dataset version to ClearML Data if opt.upload_dataset is True.
|
||||
|
||||
Arguments:
|
||||
opt (namespace) -- Commandline arguments for this run
|
||||
hyp (dict) -- Hyperparameters for this run
|
||||
|
||||
"""
|
||||
self.current_epoch = 0
|
||||
# Keep tracked of amount of logged images to enforce a limit
|
||||
self.current_epoch_logged_images = set()
|
||||
# Maximum number of images to log to clearML per epoch
|
||||
self.max_imgs_to_log_per_epoch = 16
|
||||
# Get the interval of epochs when bounding box images should be logged
|
||||
# Only for detection task though!
|
||||
if "bbox_interval" in opt:
|
||||
self.bbox_interval = opt.bbox_interval
|
||||
self.clearml = clearml
|
||||
self.task = None
|
||||
self.data_dict = None
|
||||
if self.clearml:
|
||||
self.task = Task.init(
|
||||
project_name="YOLOv5" if str(opt.project).startswith("runs/") else opt.project,
|
||||
task_name=opt.name if opt.name != "exp" else "Training",
|
||||
tags=["YOLOv5"],
|
||||
output_uri=True,
|
||||
reuse_last_task_id=opt.exist_ok,
|
||||
auto_connect_frameworks={"pytorch": False, "matplotlib": False},
|
||||
# We disconnect pytorch auto-detection, because we added manual model save points in the code
|
||||
)
|
||||
# ClearML's hooks will already grab all general parameters
|
||||
# Only the hyperparameters coming from the yaml config file
|
||||
# will have to be added manually!
|
||||
self.task.connect(hyp, name="Hyperparameters")
|
||||
self.task.connect(opt, name="Args")
|
||||
|
||||
# Make sure the code is easily remotely runnable by setting the docker image to use by the remote agent
|
||||
self.task.set_base_docker(
|
||||
"ultralytics/yolov5:latest",
|
||||
docker_arguments='--ipc=host -e="CLEARML_AGENT_SKIP_PYTHON_ENV_INSTALL=1"',
|
||||
docker_setup_bash_script="pip install clearml",
|
||||
)
|
||||
|
||||
# Get ClearML Dataset Version if requested
|
||||
if opt.data.startswith("clearml://"):
|
||||
# data_dict should have the following keys:
|
||||
# names, nc (number of classes), test, train, val (all three relative paths to ../datasets)
|
||||
self.data_dict = construct_dataset(opt.data)
|
||||
# Set data to data_dict because wandb will crash without this information and opt is the best way
|
||||
# to give it to them
|
||||
opt.data = self.data_dict
|
||||
|
||||
def log_scalars(self, metrics, epoch):
|
||||
"""
|
||||
Log scalars/metrics to ClearML.
|
||||
|
||||
Arguments:
|
||||
metrics (dict) Metrics in dict format: {"metrics/mAP": 0.8, ...}
|
||||
epoch (int) iteration number for the current set of metrics
|
||||
"""
|
||||
for k, v in metrics.items():
|
||||
title, series = k.split("/")
|
||||
self.task.get_logger().report_scalar(title, series, v, epoch)
|
||||
|
||||
def log_model(self, model_path, model_name, epoch=0):
|
||||
"""
|
||||
Log model weights to ClearML.
|
||||
|
||||
Arguments:
|
||||
model_path (PosixPath or str) Path to the model weights
|
||||
model_name (str) Name of the model visible in ClearML
|
||||
epoch (int) Iteration / epoch of the model weights
|
||||
"""
|
||||
self.task.update_output_model(
|
||||
model_path=str(model_path), name=model_name, iteration=epoch, auto_delete_file=False
|
||||
)
|
||||
|
||||
def log_summary(self, metrics):
|
||||
"""
|
||||
Log final metrics to a summary table.
|
||||
|
||||
Arguments:
|
||||
metrics (dict) Metrics in dict format: {"metrics/mAP": 0.8, ...}
|
||||
"""
|
||||
for k, v in metrics.items():
|
||||
self.task.get_logger().report_single_value(k, v)
|
||||
|
||||
def log_plot(self, title, plot_path):
|
||||
"""
|
||||
Log image as plot in the plot section of ClearML.
|
||||
|
||||
Arguments:
|
||||
title (str) Title of the plot
|
||||
plot_path (PosixPath or str) Path to the saved image file
|
||||
"""
|
||||
img = mpimg.imread(plot_path)
|
||||
fig = plt.figure()
|
||||
ax = fig.add_axes([0, 0, 1, 1], frameon=False, aspect="auto", xticks=[], yticks=[]) # no ticks
|
||||
ax.imshow(img)
|
||||
|
||||
self.task.get_logger().report_matplotlib_figure(title, "", figure=fig, report_interactive=False)
|
||||
|
||||
def log_debug_samples(self, files, title="Debug Samples"):
|
||||
"""
|
||||
Log files (images) as debug samples in the ClearML task.
|
||||
|
||||
Arguments:
|
||||
files (List(PosixPath)) a list of file paths in PosixPath format
|
||||
title (str) A title that groups together images with the same values
|
||||
"""
|
||||
for f in files:
|
||||
if f.exists():
|
||||
it = re.search(r"_batch(\d+)", f.name)
|
||||
iteration = int(it.groups()[0]) if it else 0
|
||||
self.task.get_logger().report_image(
|
||||
title=title, series=f.name.replace(f"_batch{iteration}", ""), local_path=str(f), iteration=iteration
|
||||
)
|
||||
|
||||
def log_image_with_boxes(self, image_path, boxes, class_names, image, conf_threshold=0.25):
|
||||
"""
|
||||
Draw the bounding boxes on a single image and report the result as a ClearML debug sample.
|
||||
|
||||
Arguments:
|
||||
image_path (PosixPath) the path the original image file
|
||||
boxes (list): list of scaled predictions in the format - [xmin, ymin, xmax, ymax, confidence, class]
|
||||
class_names (dict): dict containing mapping of class int to class name
|
||||
image (Tensor): A torch tensor containing the actual image data
|
||||
"""
|
||||
if (
|
||||
len(self.current_epoch_logged_images) < self.max_imgs_to_log_per_epoch
|
||||
and self.current_epoch >= 0
|
||||
and (self.current_epoch % self.bbox_interval == 0 and image_path not in self.current_epoch_logged_images)
|
||||
):
|
||||
im = np.ascontiguousarray(np.moveaxis(image.mul(255).clamp(0, 255).byte().cpu().numpy(), 0, 2))
|
||||
annotator = Annotator(im=im, pil=True)
|
||||
for i, (conf, class_nr, box) in enumerate(zip(boxes[:, 4], boxes[:, 5], boxes[:, :4])):
|
||||
color = colors(i)
|
||||
|
||||
class_name = class_names[int(class_nr)]
|
||||
confidence_percentage = round(float(conf) * 100, 2)
|
||||
label = f"{class_name}: {confidence_percentage}%"
|
||||
|
||||
if conf > conf_threshold:
|
||||
annotator.rectangle(box.cpu().numpy(), outline=color)
|
||||
annotator.box_label(box.cpu().numpy(), label=label, color=color)
|
||||
|
||||
annotated_image = annotator.result()
|
||||
self.task.get_logger().report_image(
|
||||
title="Bounding Boxes", series=image_path.name, iteration=self.current_epoch, image=annotated_image
|
||||
)
|
||||
self.current_epoch_logged_images.add(image_path)
|
||||
@@ -1,90 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
from clearml import Task
|
||||
|
||||
# Connecting ClearML with the current process,
|
||||
# from here on everything is logged automatically
|
||||
from clearml.automation import HyperParameterOptimizer, UniformParameterRange
|
||||
from clearml.automation.optuna import OptimizerOptuna
|
||||
|
||||
task = Task.init(
|
||||
project_name="Hyper-Parameter Optimization",
|
||||
task_name="YOLOv5",
|
||||
task_type=Task.TaskTypes.optimizer,
|
||||
reuse_last_task_id=False,
|
||||
)
|
||||
|
||||
# Example use case:
|
||||
optimizer = HyperParameterOptimizer(
|
||||
# This is the experiment we want to optimize
|
||||
base_task_id="<your_template_task_id>",
|
||||
# here we define the hyper-parameters to optimize
|
||||
# Notice: The parameter name should exactly match what you see in the UI: <section_name>/<parameter>
|
||||
# For Example, here we see in the base experiment a section Named: "General"
|
||||
# under it a parameter named "batch_size", this becomes "General/batch_size"
|
||||
# If you have `argparse` for example, then arguments will appear under the "Args" section,
|
||||
# and you should instead pass "Args/batch_size"
|
||||
hyper_parameters=[
|
||||
UniformParameterRange("Hyperparameters/lr0", min_value=1e-5, max_value=1e-1),
|
||||
UniformParameterRange("Hyperparameters/lrf", min_value=0.01, max_value=1.0),
|
||||
UniformParameterRange("Hyperparameters/momentum", min_value=0.6, max_value=0.98),
|
||||
UniformParameterRange("Hyperparameters/weight_decay", min_value=0.0, max_value=0.001),
|
||||
UniformParameterRange("Hyperparameters/warmup_epochs", min_value=0.0, max_value=5.0),
|
||||
UniformParameterRange("Hyperparameters/warmup_momentum", min_value=0.0, max_value=0.95),
|
||||
UniformParameterRange("Hyperparameters/warmup_bias_lr", min_value=0.0, max_value=0.2),
|
||||
UniformParameterRange("Hyperparameters/box", min_value=0.02, max_value=0.2),
|
||||
UniformParameterRange("Hyperparameters/cls", min_value=0.2, max_value=4.0),
|
||||
UniformParameterRange("Hyperparameters/cls_pw", min_value=0.5, max_value=2.0),
|
||||
UniformParameterRange("Hyperparameters/obj", min_value=0.2, max_value=4.0),
|
||||
UniformParameterRange("Hyperparameters/obj_pw", min_value=0.5, max_value=2.0),
|
||||
UniformParameterRange("Hyperparameters/iou_t", min_value=0.1, max_value=0.7),
|
||||
UniformParameterRange("Hyperparameters/anchor_t", min_value=2.0, max_value=8.0),
|
||||
UniformParameterRange("Hyperparameters/fl_gamma", min_value=0.0, max_value=4.0),
|
||||
UniformParameterRange("Hyperparameters/hsv_h", min_value=0.0, max_value=0.1),
|
||||
UniformParameterRange("Hyperparameters/hsv_s", min_value=0.0, max_value=0.9),
|
||||
UniformParameterRange("Hyperparameters/hsv_v", min_value=0.0, max_value=0.9),
|
||||
UniformParameterRange("Hyperparameters/degrees", min_value=0.0, max_value=45.0),
|
||||
UniformParameterRange("Hyperparameters/translate", min_value=0.0, max_value=0.9),
|
||||
UniformParameterRange("Hyperparameters/scale", min_value=0.0, max_value=0.9),
|
||||
UniformParameterRange("Hyperparameters/shear", min_value=0.0, max_value=10.0),
|
||||
UniformParameterRange("Hyperparameters/perspective", min_value=0.0, max_value=0.001),
|
||||
UniformParameterRange("Hyperparameters/flipud", min_value=0.0, max_value=1.0),
|
||||
UniformParameterRange("Hyperparameters/fliplr", min_value=0.0, max_value=1.0),
|
||||
UniformParameterRange("Hyperparameters/mosaic", min_value=0.0, max_value=1.0),
|
||||
UniformParameterRange("Hyperparameters/mixup", min_value=0.0, max_value=1.0),
|
||||
UniformParameterRange("Hyperparameters/copy_paste", min_value=0.0, max_value=1.0),
|
||||
],
|
||||
# this is the objective metric we want to maximize/minimize
|
||||
objective_metric_title="metrics",
|
||||
objective_metric_series="mAP_0.5",
|
||||
# now we decide if we want to maximize it or minimize it (accuracy we maximize)
|
||||
objective_metric_sign="max",
|
||||
# let us limit the number of concurrent experiments,
|
||||
# this in turn will make sure we don't bombard the scheduler with experiments.
|
||||
# if we have an auto-scaler connected, this, by proxy, will limit the number of machine
|
||||
max_number_of_concurrent_tasks=1,
|
||||
# this is the optimizer class (actually doing the optimization)
|
||||
# Currently, we can choose from GridSearch, RandomSearch or OptimizerBOHB (Bayesian optimization Hyper-Band)
|
||||
optimizer_class=OptimizerOptuna,
|
||||
# If specified only the top K performing Tasks will be kept, the others will be automatically archived
|
||||
save_top_k_tasks_only=5, # 5,
|
||||
compute_time_limit=None,
|
||||
total_max_jobs=20,
|
||||
min_iteration_per_job=None,
|
||||
max_iteration_per_job=None,
|
||||
)
|
||||
|
||||
# report every 10 seconds, this is way too often, but we are testing here
|
||||
optimizer.set_report_period(10 / 60)
|
||||
# You can also use the line below instead to run all the optimizer tasks locally, without using queues or agent
|
||||
# an_optimizer.start_locally(job_complete_callback=job_complete_callback)
|
||||
# set the time limit for the optimization process (2 hours)
|
||||
optimizer.set_time_limit(in_minutes=120.0)
|
||||
# Start the optimization process in the local environment
|
||||
optimizer.start_locally()
|
||||
# wait until process is done (notice we are controlling the optimization process in the background)
|
||||
optimizer.wait()
|
||||
# make sure background optimization stopped
|
||||
optimizer.stop()
|
||||
|
||||
print("We are done, good bye")
|
||||
@@ -1,250 +0,0 @@
|
||||
<img src="https://cdn.comet.ml/img/notebook_logo.png">
|
||||
|
||||
# YOLOv5 with Comet
|
||||
|
||||
This guide will cover how to use YOLOv5 with [Comet](https://bit.ly/yolov5-readme-comet2)
|
||||
|
||||
# About Comet
|
||||
|
||||
Comet builds tools that help data scientists, engineers, and team leaders accelerate and optimize machine learning and deep learning models.
|
||||
|
||||
Track and visualize model metrics in real time, save your hyperparameters, datasets, and model checkpoints, and visualize your model predictions with [Comet Custom Panels](https://www.comet.com/docs/v2/guides/comet-dashboard/code-panels/about-panels/?utm_source=yolov5&utm_medium=partner&utm_campaign=partner_yolov5_2022&utm_content=github)! Comet makes sure you never lose track of your work and makes it easy to share results and collaborate across teams of all sizes!
|
||||
|
||||
# Getting Started
|
||||
|
||||
## Install Comet
|
||||
|
||||
```shell
|
||||
pip install comet_ml
|
||||
```
|
||||
|
||||
## Configure Comet Credentials
|
||||
|
||||
There are two ways to configure Comet with YOLOv5.
|
||||
|
||||
You can either set your credentials through environment variables
|
||||
|
||||
**Environment Variables**
|
||||
|
||||
```shell
|
||||
export COMET_API_KEY=<Your Comet API Key>
|
||||
export COMET_PROJECT_NAME=<Your Comet Project Name> # This will default to 'yolov5'
|
||||
```
|
||||
|
||||
Or create a `.comet.config` file in your working directory and set your credentials there.
|
||||
|
||||
**Comet Configuration File**
|
||||
|
||||
```
|
||||
[comet]
|
||||
api_key=<Your Comet API Key>
|
||||
project_name=<Your Comet Project Name> # This will default to 'yolov5'
|
||||
```
|
||||
|
||||
## Run the Training Script
|
||||
|
||||
```shell
|
||||
# Train YOLOv5s on COCO128 for 5 epochs
|
||||
python train.py --img 640 --batch 16 --epochs 5 --data coco128.yaml --weights yolov5s.pt
|
||||
```
|
||||
|
||||
That's it! Comet will automatically log your hyperparameters, command line arguments, training and validation metrics. You can visualize and analyze your runs in the Comet UI
|
||||
|
||||
<img width="1920" alt="yolo-ui" src="https://user-images.githubusercontent.com/26833433/202851203-164e94e1-2238-46dd-91f8-de020e9d6b41.png">
|
||||
|
||||
# Try out an Example!
|
||||
|
||||
Check out an example of a [completed run here](https://www.comet.com/examples/comet-example-yolov5/a0e29e0e9b984e4a822db2a62d0cb357?experiment-tab=chart&showOutliers=true&smoothing=0&transformY=smoothing&xAxis=step&utm_source=yolov5&utm_medium=partner&utm_campaign=partner_yolov5_2022&utm_content=github)
|
||||
|
||||
Or better yet, try it out yourself in this Colab Notebook
|
||||
|
||||
[](https://colab.research.google.com/github/comet-ml/comet-examples/blob/master/integrations/model-training/yolov5/notebooks/Comet_and_YOLOv5.ipynb)
|
||||
|
||||
# Log automatically
|
||||
|
||||
By default, Comet will log the following items
|
||||
|
||||
## Metrics
|
||||
|
||||
- Box Loss, Object Loss, Classification Loss for the training and validation data
|
||||
- mAP_0.5, mAP_0.5:0.95 metrics for the validation data.
|
||||
- Precision and Recall for the validation data
|
||||
|
||||
## Parameters
|
||||
|
||||
- Model Hyperparameters
|
||||
- All parameters passed through the command line options
|
||||
|
||||
## Visualizations
|
||||
|
||||
- Confusion Matrix of the model predictions on the validation data
|
||||
- Plots for the PR and F1 curves across all classes
|
||||
- Correlogram of the Class Labels
|
||||
|
||||
# Configure Comet Logging
|
||||
|
||||
Comet can be configured to log additional data either through command line flags passed to the training script or through environment variables.
|
||||
|
||||
```shell
|
||||
export COMET_MODE=online # Set whether to run Comet in 'online' or 'offline' mode. Defaults to online
|
||||
export COMET_MODEL_NAME=<your model name> #Set the name for the saved model. Defaults to yolov5
|
||||
export COMET_LOG_CONFUSION_MATRIX=false # Set to disable logging a Comet Confusion Matrix. Defaults to true
|
||||
export COMET_MAX_IMAGE_UPLOADS=<number of allowed images to upload to Comet> # Controls how many total image predictions to log to Comet. Defaults to 100.
|
||||
export COMET_LOG_PER_CLASS_METRICS=true # Set to log evaluation metrics for each detected class at the end of training. Defaults to false
|
||||
export COMET_DEFAULT_CHECKPOINT_FILENAME=<your checkpoint filename> # Set this if you would like to resume training from a different checkpoint. Defaults to 'last.pt'
|
||||
export COMET_LOG_BATCH_LEVEL_METRICS=true # Set this if you would like to log training metrics at the batch level. Defaults to false.
|
||||
export COMET_LOG_PREDICTIONS=true # Set this to false to disable logging model predictions
|
||||
```
|
||||
|
||||
## Logging Checkpoints with Comet
|
||||
|
||||
Logging Models to Comet is disabled by default. To enable it, pass the `save-period` argument to the training script. This will save the logged checkpoints to Comet based on the interval value provided by `save-period`
|
||||
|
||||
```shell
|
||||
python train.py \
|
||||
--img 640 \
|
||||
--batch 16 \
|
||||
--epochs 5 \
|
||||
--data coco128.yaml \
|
||||
--weights yolov5s.pt \
|
||||
--save-period 1
|
||||
```
|
||||
|
||||
## Logging Model Predictions
|
||||
|
||||
By default, model predictions (images, ground truth labels and bounding boxes) will be logged to Comet.
|
||||
|
||||
You can control the frequency of logged predictions and the associated images by passing the `bbox_interval` command line argument. Predictions can be visualized using Comet's Object Detection Custom Panel. This frequency corresponds to every Nth batch of data per epoch. In the example below, we are logging every 2nd batch of data for each epoch.
|
||||
|
||||
**Note:** The YOLOv5 validation dataloader will default to a batch size of 32, so you will have to set the logging frequency accordingly.
|
||||
|
||||
Here is an [example project using the Panel](https://www.comet.com/examples/comet-example-yolov5?shareable=YcwMiJaZSXfcEXpGOHDD12vA1&utm_source=yolov5&utm_medium=partner&utm_campaign=partner_yolov5_2022&utm_content=github)
|
||||
|
||||
```shell
|
||||
python train.py \
|
||||
--img 640 \
|
||||
--batch 16 \
|
||||
--epochs 5 \
|
||||
--data coco128.yaml \
|
||||
--weights yolov5s.pt \
|
||||
--bbox_interval 2
|
||||
```
|
||||
|
||||
### Controlling the number of Prediction Images logged to Comet
|
||||
|
||||
When logging predictions from YOLOv5, Comet will log the images associated with each set of predictions. By default a maximum of 100 validation images are logged. You can increase or decrease this number using the `COMET_MAX_IMAGE_UPLOADS` environment variable.
|
||||
|
||||
```shell
|
||||
env COMET_MAX_IMAGE_UPLOADS=200 python train.py \
|
||||
--img 640 \
|
||||
--batch 16 \
|
||||
--epochs 5 \
|
||||
--data coco128.yaml \
|
||||
--weights yolov5s.pt \
|
||||
--bbox_interval 1
|
||||
```
|
||||
|
||||
### Logging Class Level Metrics
|
||||
|
||||
Use the `COMET_LOG_PER_CLASS_METRICS` environment variable to log mAP, precision, recall, f1 for each class.
|
||||
|
||||
```shell
|
||||
env COMET_LOG_PER_CLASS_METRICS=true python train.py \
|
||||
--img 640 \
|
||||
--batch 16 \
|
||||
--epochs 5 \
|
||||
--data coco128.yaml \
|
||||
--weights yolov5s.pt
|
||||
```
|
||||
|
||||
## Uploading a Dataset to Comet Artifacts
|
||||
|
||||
If you would like to store your data using [Comet Artifacts](https://www.comet.com/docs/v2/guides/data-management/using-artifacts/#learn-more?utm_source=yolov5&utm_medium=partner&utm_campaign=partner_yolov5_2022&utm_content=github), you can do so using the `upload_dataset` flag.
|
||||
|
||||
The dataset be organized in the way described in the [YOLOv5 documentation](https://docs.ultralytics.com/yolov5/tutorials/train_custom_data/). The dataset config `yaml` file must follow the same format as that of the `coco128.yaml` file.
|
||||
|
||||
```shell
|
||||
python train.py \
|
||||
--img 640 \
|
||||
--batch 16 \
|
||||
--epochs 5 \
|
||||
--data coco128.yaml \
|
||||
--weights yolov5s.pt \
|
||||
--upload_dataset
|
||||
```
|
||||
|
||||
You can find the uploaded dataset in the Artifacts tab in your Comet Workspace <img width="1073" alt="artifact-1" src="https://user-images.githubusercontent.com/7529846/186929193-162718bf-ec7b-4eb9-8c3b-86b3763ef8ea.png">
|
||||
|
||||
You can preview the data directly in the Comet UI. <img width="1082" alt="artifact-2" src="https://user-images.githubusercontent.com/7529846/186929215-432c36a9-c109-4eb0-944b-84c2786590d6.png">
|
||||
|
||||
Artifacts are versioned and also support adding metadata about the dataset. Comet will automatically log the metadata from your dataset `yaml` file <img width="963" alt="artifact-3" src="https://user-images.githubusercontent.com/7529846/186929256-9d44d6eb-1a19-42de-889a-bcbca3018f2e.png">
|
||||
|
||||
### Using a saved Artifact
|
||||
|
||||
If you would like to use a dataset from Comet Artifacts, set the `path` variable in your dataset `yaml` file to point to the following Artifact resource URL.
|
||||
|
||||
```
|
||||
# contents of artifact.yaml file
|
||||
path: "comet://<workspace name>/<artifact name>:<artifact version or alias>"
|
||||
```
|
||||
|
||||
Then pass this file to your training script in the following way
|
||||
|
||||
```shell
|
||||
python train.py \
|
||||
--img 640 \
|
||||
--batch 16 \
|
||||
--epochs 5 \
|
||||
--data artifact.yaml \
|
||||
--weights yolov5s.pt
|
||||
```
|
||||
|
||||
Artifacts also allow you to track the lineage of data as it flows through your Experimentation workflow. Here you can see a graph that shows you all the experiments that have used your uploaded dataset. <img width="1391" alt="artifact-4" src="https://user-images.githubusercontent.com/7529846/186929264-4c4014fa-fe51-4f3c-a5c5-f6d24649b1b4.png">
|
||||
|
||||
## Resuming a Training Run
|
||||
|
||||
If your training run is interrupted for any reason, e.g. disrupted internet connection, you can resume the run using the `resume` flag and the Comet Run Path.
|
||||
|
||||
The Run Path has the following format `comet://<your workspace name>/<your project name>/<experiment id>`.
|
||||
|
||||
This will restore the run to its state before the interruption, which includes restoring the model from a checkpoint, restoring all hyperparameters and training arguments and downloading Comet dataset Artifacts if they were used in the original run. The resumed run will continue logging to the existing Experiment in the Comet UI
|
||||
|
||||
```shell
|
||||
python train.py \
|
||||
--resume "comet://<your run path>"
|
||||
```
|
||||
|
||||
## Hyperparameter Search with the Comet Optimizer
|
||||
|
||||
YOLOv5 is also integrated with Comet's Optimizer, making is simple to visualize hyperparameter sweeps in the Comet UI.
|
||||
|
||||
### Configuring an Optimizer Sweep
|
||||
|
||||
To configure the Comet Optimizer, you will have to create a JSON file with the information about the sweep. An example file has been provided in `utils/loggers/comet/optimizer_config.json`
|
||||
|
||||
```shell
|
||||
python utils/loggers/comet/hpo.py \
|
||||
--comet_optimizer_config "utils/loggers/comet/optimizer_config.json"
|
||||
```
|
||||
|
||||
The `hpo.py` script accepts the same arguments as `train.py`. If you wish to pass additional arguments to your sweep simply add them after the script.
|
||||
|
||||
```shell
|
||||
python utils/loggers/comet/hpo.py \
|
||||
--comet_optimizer_config "utils/loggers/comet/optimizer_config.json" \
|
||||
--save-period 1 \
|
||||
--bbox_interval 1
|
||||
```
|
||||
|
||||
### Running a Sweep in Parallel
|
||||
|
||||
```shell
|
||||
comet optimizer -j <set number of workers> utils/loggers/comet/hpo.py \
|
||||
utils/loggers/comet/optimizer_config.json"
|
||||
```
|
||||
|
||||
### Visualizing Results
|
||||
|
||||
Comet provides a number of ways to visualize the results of your sweep. Take a look at a [project with a completed sweep here](https://www.comet.com/examples/comet-example-yolov5/view/PrlArHGuuhDTKC1UuBmTtOSXD/panels?utm_source=yolov5&utm_medium=partner&utm_campaign=partner_yolov5_2022&utm_content=github)
|
||||
|
||||
<img width="1626" alt="hyperparameter-yolo" src="https://user-images.githubusercontent.com/7529846/186914869-7dc1de14-583f-4323-967b-c9a66a29e495.png">
|
||||
@@ -1,549 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
import glob
|
||||
import json
|
||||
import logging
|
||||
import os
|
||||
import sys
|
||||
from pathlib import Path
|
||||
|
||||
logger = logging.getLogger(__name__)
|
||||
|
||||
FILE = Path(__file__).resolve()
|
||||
ROOT = FILE.parents[3] # YOLOv5 root directory
|
||||
if str(ROOT) not in sys.path:
|
||||
sys.path.append(str(ROOT)) # add ROOT to PATH
|
||||
|
||||
try:
|
||||
import comet_ml
|
||||
|
||||
# Project Configuration
|
||||
config = comet_ml.config.get_config()
|
||||
COMET_PROJECT_NAME = config.get_string(os.getenv("COMET_PROJECT_NAME"), "comet.project_name", default="yolov5")
|
||||
except ImportError:
|
||||
comet_ml = None
|
||||
COMET_PROJECT_NAME = None
|
||||
|
||||
import PIL
|
||||
import torch
|
||||
import torchvision.transforms as T
|
||||
import yaml
|
||||
|
||||
from utils.dataloaders import img2label_paths
|
||||
from utils.general import check_dataset, scale_boxes, xywh2xyxy
|
||||
from utils.metrics import box_iou
|
||||
|
||||
COMET_PREFIX = "comet://"
|
||||
|
||||
COMET_MODE = os.getenv("COMET_MODE", "online")
|
||||
|
||||
# Model Saving Settings
|
||||
COMET_MODEL_NAME = os.getenv("COMET_MODEL_NAME", "yolov5")
|
||||
|
||||
# Dataset Artifact Settings
|
||||
COMET_UPLOAD_DATASET = os.getenv("COMET_UPLOAD_DATASET", "false").lower() == "true"
|
||||
|
||||
# Evaluation Settings
|
||||
COMET_LOG_CONFUSION_MATRIX = os.getenv("COMET_LOG_CONFUSION_MATRIX", "true").lower() == "true"
|
||||
COMET_LOG_PREDICTIONS = os.getenv("COMET_LOG_PREDICTIONS", "true").lower() == "true"
|
||||
COMET_MAX_IMAGE_UPLOADS = int(os.getenv("COMET_MAX_IMAGE_UPLOADS", 100))
|
||||
|
||||
# Confusion Matrix Settings
|
||||
CONF_THRES = float(os.getenv("CONF_THRES", 0.001))
|
||||
IOU_THRES = float(os.getenv("IOU_THRES", 0.6))
|
||||
|
||||
# Batch Logging Settings
|
||||
COMET_LOG_BATCH_METRICS = os.getenv("COMET_LOG_BATCH_METRICS", "false").lower() == "true"
|
||||
COMET_BATCH_LOGGING_INTERVAL = os.getenv("COMET_BATCH_LOGGING_INTERVAL", 1)
|
||||
COMET_PREDICTION_LOGGING_INTERVAL = os.getenv("COMET_PREDICTION_LOGGING_INTERVAL", 1)
|
||||
COMET_LOG_PER_CLASS_METRICS = os.getenv("COMET_LOG_PER_CLASS_METRICS", "false").lower() == "true"
|
||||
|
||||
RANK = int(os.getenv("RANK", -1))
|
||||
|
||||
to_pil = T.ToPILImage()
|
||||
|
||||
|
||||
class CometLogger:
|
||||
"""Log metrics, parameters, source code, models and much more with Comet."""
|
||||
|
||||
def __init__(self, opt, hyp, run_id=None, job_type="Training", **experiment_kwargs) -> None:
|
||||
"""Initializes CometLogger with given options, hyperparameters, run ID, job type, and additional experiment
|
||||
arguments.
|
||||
"""
|
||||
self.job_type = job_type
|
||||
self.opt = opt
|
||||
self.hyp = hyp
|
||||
|
||||
# Comet Flags
|
||||
self.comet_mode = COMET_MODE
|
||||
|
||||
self.save_model = opt.save_period > -1
|
||||
self.model_name = COMET_MODEL_NAME
|
||||
|
||||
# Batch Logging Settings
|
||||
self.log_batch_metrics = COMET_LOG_BATCH_METRICS
|
||||
self.comet_log_batch_interval = COMET_BATCH_LOGGING_INTERVAL
|
||||
|
||||
# Dataset Artifact Settings
|
||||
self.upload_dataset = self.opt.upload_dataset or COMET_UPLOAD_DATASET
|
||||
self.resume = self.opt.resume
|
||||
|
||||
self.default_experiment_kwargs = {
|
||||
"log_code": False,
|
||||
"log_env_gpu": True,
|
||||
"log_env_cpu": True,
|
||||
"project_name": COMET_PROJECT_NAME,
|
||||
} | experiment_kwargs
|
||||
self.experiment = self._get_experiment(self.comet_mode, run_id)
|
||||
self.experiment.set_name(self.opt.name)
|
||||
|
||||
self.data_dict = self.check_dataset(self.opt.data)
|
||||
self.class_names = self.data_dict["names"]
|
||||
self.num_classes = self.data_dict["nc"]
|
||||
|
||||
self.logged_images_count = 0
|
||||
self.max_images = COMET_MAX_IMAGE_UPLOADS
|
||||
|
||||
if run_id is None:
|
||||
self.experiment.log_other("Created from", "YOLOv5")
|
||||
if not isinstance(self.experiment, comet_ml.OfflineExperiment):
|
||||
workspace, project_name, experiment_id = self.experiment.url.split("/")[-3:]
|
||||
self.experiment.log_other(
|
||||
"Run Path",
|
||||
f"{workspace}/{project_name}/{experiment_id}",
|
||||
)
|
||||
self.log_parameters(vars(opt))
|
||||
self.log_parameters(self.opt.hyp)
|
||||
self.log_asset_data(
|
||||
self.opt.hyp,
|
||||
name="hyperparameters.json",
|
||||
metadata={"type": "hyp-config-file"},
|
||||
)
|
||||
self.log_asset(
|
||||
f"{self.opt.save_dir}/opt.yaml",
|
||||
metadata={"type": "opt-config-file"},
|
||||
)
|
||||
|
||||
self.comet_log_confusion_matrix = COMET_LOG_CONFUSION_MATRIX
|
||||
|
||||
if hasattr(self.opt, "conf_thres"):
|
||||
self.conf_thres = self.opt.conf_thres
|
||||
else:
|
||||
self.conf_thres = CONF_THRES
|
||||
if hasattr(self.opt, "iou_thres"):
|
||||
self.iou_thres = self.opt.iou_thres
|
||||
else:
|
||||
self.iou_thres = IOU_THRES
|
||||
|
||||
self.log_parameters({"val_iou_threshold": self.iou_thres, "val_conf_threshold": self.conf_thres})
|
||||
|
||||
self.comet_log_predictions = COMET_LOG_PREDICTIONS
|
||||
if self.opt.bbox_interval == -1:
|
||||
self.comet_log_prediction_interval = 1 if self.opt.epochs < 10 else self.opt.epochs // 10
|
||||
else:
|
||||
self.comet_log_prediction_interval = self.opt.bbox_interval
|
||||
|
||||
if self.comet_log_predictions:
|
||||
self.metadata_dict = {}
|
||||
self.logged_image_names = []
|
||||
|
||||
self.comet_log_per_class_metrics = COMET_LOG_PER_CLASS_METRICS
|
||||
|
||||
self.experiment.log_others(
|
||||
{
|
||||
"comet_mode": COMET_MODE,
|
||||
"comet_max_image_uploads": COMET_MAX_IMAGE_UPLOADS,
|
||||
"comet_log_per_class_metrics": COMET_LOG_PER_CLASS_METRICS,
|
||||
"comet_log_batch_metrics": COMET_LOG_BATCH_METRICS,
|
||||
"comet_log_confusion_matrix": COMET_LOG_CONFUSION_MATRIX,
|
||||
"comet_model_name": COMET_MODEL_NAME,
|
||||
}
|
||||
)
|
||||
|
||||
# Check if running the Experiment with the Comet Optimizer
|
||||
if hasattr(self.opt, "comet_optimizer_id"):
|
||||
self.experiment.log_other("optimizer_id", self.opt.comet_optimizer_id)
|
||||
self.experiment.log_other("optimizer_objective", self.opt.comet_optimizer_objective)
|
||||
self.experiment.log_other("optimizer_metric", self.opt.comet_optimizer_metric)
|
||||
self.experiment.log_other("optimizer_parameters", json.dumps(self.hyp))
|
||||
|
||||
def _get_experiment(self, mode, experiment_id=None):
|
||||
"""Returns a new or existing Comet.ml experiment based on mode and optional experiment_id."""
|
||||
if mode == "offline":
|
||||
return (
|
||||
comet_ml.ExistingOfflineExperiment(
|
||||
previous_experiment=experiment_id,
|
||||
**self.default_experiment_kwargs,
|
||||
)
|
||||
if experiment_id is not None
|
||||
else comet_ml.OfflineExperiment(
|
||||
**self.default_experiment_kwargs,
|
||||
)
|
||||
)
|
||||
try:
|
||||
if experiment_id is not None:
|
||||
return comet_ml.ExistingExperiment(
|
||||
previous_experiment=experiment_id,
|
||||
**self.default_experiment_kwargs,
|
||||
)
|
||||
|
||||
return comet_ml.Experiment(**self.default_experiment_kwargs)
|
||||
|
||||
except ValueError:
|
||||
logger.warning(
|
||||
"COMET WARNING: "
|
||||
"Comet credentials have not been set. "
|
||||
"Comet will default to offline logging. "
|
||||
"Please set your credentials to enable online logging."
|
||||
)
|
||||
return self._get_experiment("offline", experiment_id)
|
||||
|
||||
return
|
||||
|
||||
def log_metrics(self, log_dict, **kwargs):
|
||||
"""Logs metrics to the current experiment, accepting a dictionary of metric names and values."""
|
||||
self.experiment.log_metrics(log_dict, **kwargs)
|
||||
|
||||
def log_parameters(self, log_dict, **kwargs):
|
||||
"""Logs parameters to the current experiment, accepting a dictionary of parameter names and values."""
|
||||
self.experiment.log_parameters(log_dict, **kwargs)
|
||||
|
||||
def log_asset(self, asset_path, **kwargs):
|
||||
"""Logs a file or directory as an asset to the current experiment."""
|
||||
self.experiment.log_asset(asset_path, **kwargs)
|
||||
|
||||
def log_asset_data(self, asset, **kwargs):
|
||||
"""Logs in-memory data as an asset to the current experiment, with optional kwargs."""
|
||||
self.experiment.log_asset_data(asset, **kwargs)
|
||||
|
||||
def log_image(self, img, **kwargs):
|
||||
"""Logs an image to the current experiment with optional kwargs."""
|
||||
self.experiment.log_image(img, **kwargs)
|
||||
|
||||
def log_model(self, path, opt, epoch, fitness_score, best_model=False):
|
||||
"""Logs model checkpoint to experiment with path, options, epoch, fitness, and best model flag."""
|
||||
if not self.save_model:
|
||||
return
|
||||
|
||||
model_metadata = {
|
||||
"fitness_score": fitness_score[-1],
|
||||
"epochs_trained": epoch + 1,
|
||||
"save_period": opt.save_period,
|
||||
"total_epochs": opt.epochs,
|
||||
}
|
||||
|
||||
model_files = glob.glob(f"{path}/*.pt")
|
||||
for model_path in model_files:
|
||||
name = Path(model_path).name
|
||||
|
||||
self.experiment.log_model(
|
||||
self.model_name,
|
||||
file_or_folder=model_path,
|
||||
file_name=name,
|
||||
metadata=model_metadata,
|
||||
overwrite=True,
|
||||
)
|
||||
|
||||
def check_dataset(self, data_file):
|
||||
"""Validates the dataset configuration by loading the YAML file specified in `data_file`."""
|
||||
with open(data_file) as f:
|
||||
data_config = yaml.safe_load(f)
|
||||
|
||||
path = data_config.get("path")
|
||||
if path and path.startswith(COMET_PREFIX):
|
||||
path = data_config["path"].replace(COMET_PREFIX, "")
|
||||
return self.download_dataset_artifact(path)
|
||||
self.log_asset(self.opt.data, metadata={"type": "data-config-file"})
|
||||
|
||||
return check_dataset(data_file)
|
||||
|
||||
def log_predictions(self, image, labelsn, path, shape, predn):
|
||||
"""Logs predictions with IOU filtering, given image, labels, path, shape, and predictions."""
|
||||
if self.logged_images_count >= self.max_images:
|
||||
return
|
||||
detections = predn[predn[:, 4] > self.conf_thres]
|
||||
iou = box_iou(labelsn[:, 1:], detections[:, :4])
|
||||
mask, _ = torch.where(iou > self.iou_thres)
|
||||
if len(mask) == 0:
|
||||
return
|
||||
|
||||
filtered_detections = detections[mask]
|
||||
filtered_labels = labelsn[mask]
|
||||
|
||||
image_id = path.split("/")[-1].split(".")[0]
|
||||
image_name = f"{image_id}_curr_epoch_{self.experiment.curr_epoch}"
|
||||
if image_name not in self.logged_image_names:
|
||||
native_scale_image = PIL.Image.open(path)
|
||||
self.log_image(native_scale_image, name=image_name)
|
||||
self.logged_image_names.append(image_name)
|
||||
|
||||
metadata = [
|
||||
{
|
||||
"label": f"{self.class_names[int(cls)]}-gt",
|
||||
"score": 100,
|
||||
"box": {"x": xyxy[0], "y": xyxy[1], "x2": xyxy[2], "y2": xyxy[3]},
|
||||
}
|
||||
for cls, *xyxy in filtered_labels.tolist()
|
||||
]
|
||||
metadata.extend(
|
||||
{
|
||||
"label": f"{self.class_names[int(cls)]}",
|
||||
"score": conf * 100,
|
||||
"box": {"x": xyxy[0], "y": xyxy[1], "x2": xyxy[2], "y2": xyxy[3]},
|
||||
}
|
||||
for *xyxy, conf, cls in filtered_detections.tolist()
|
||||
)
|
||||
self.metadata_dict[image_name] = metadata
|
||||
self.logged_images_count += 1
|
||||
|
||||
return
|
||||
|
||||
def preprocess_prediction(self, image, labels, shape, pred):
|
||||
"""Processes prediction data, resizing labels and adding dataset metadata."""
|
||||
nl, _ = labels.shape[0], pred.shape[0]
|
||||
|
||||
# Predictions
|
||||
if self.opt.single_cls:
|
||||
pred[:, 5] = 0
|
||||
|
||||
predn = pred.clone()
|
||||
scale_boxes(image.shape[1:], predn[:, :4], shape[0], shape[1])
|
||||
|
||||
labelsn = None
|
||||
if nl:
|
||||
tbox = xywh2xyxy(labels[:, 1:5]) # target boxes
|
||||
scale_boxes(image.shape[1:], tbox, shape[0], shape[1]) # native-space labels
|
||||
labelsn = torch.cat((labels[:, 0:1], tbox), 1) # native-space labels
|
||||
scale_boxes(image.shape[1:], predn[:, :4], shape[0], shape[1]) # native-space pred
|
||||
|
||||
return predn, labelsn
|
||||
|
||||
def add_assets_to_artifact(self, artifact, path, asset_path, split):
|
||||
"""Adds image and label assets to a wandb artifact given dataset split and paths."""
|
||||
img_paths = sorted(glob.glob(f"{asset_path}/*"))
|
||||
label_paths = img2label_paths(img_paths)
|
||||
|
||||
for image_file, label_file in zip(img_paths, label_paths):
|
||||
image_logical_path, label_logical_path = map(lambda x: os.path.relpath(x, path), [image_file, label_file])
|
||||
|
||||
try:
|
||||
artifact.add(
|
||||
image_file,
|
||||
logical_path=image_logical_path,
|
||||
metadata={"split": split},
|
||||
)
|
||||
artifact.add(
|
||||
label_file,
|
||||
logical_path=label_logical_path,
|
||||
metadata={"split": split},
|
||||
)
|
||||
except ValueError as e:
|
||||
logger.error("COMET ERROR: Error adding file to Artifact. Skipping file.")
|
||||
logger.error(f"COMET ERROR: {e}")
|
||||
continue
|
||||
|
||||
return artifact
|
||||
|
||||
def upload_dataset_artifact(self):
|
||||
"""Uploads a YOLOv5 dataset as an artifact to the Comet.ml platform."""
|
||||
dataset_name = self.data_dict.get("dataset_name", "yolov5-dataset")
|
||||
path = str((ROOT / Path(self.data_dict["path"])).resolve())
|
||||
|
||||
metadata = self.data_dict.copy()
|
||||
for key in ["train", "val", "test"]:
|
||||
split_path = metadata.get(key)
|
||||
if split_path is not None:
|
||||
metadata[key] = split_path.replace(path, "")
|
||||
|
||||
artifact = comet_ml.Artifact(name=dataset_name, artifact_type="dataset", metadata=metadata)
|
||||
for key in metadata.keys():
|
||||
if key in ["train", "val", "test"]:
|
||||
if isinstance(self.upload_dataset, str) and (key != self.upload_dataset):
|
||||
continue
|
||||
|
||||
asset_path = self.data_dict.get(key)
|
||||
if asset_path is not None:
|
||||
artifact = self.add_assets_to_artifact(artifact, path, asset_path, key)
|
||||
|
||||
self.experiment.log_artifact(artifact)
|
||||
|
||||
return
|
||||
|
||||
def download_dataset_artifact(self, artifact_path):
|
||||
"""Downloads a dataset artifact to a specified directory using the experiment's logged artifact."""
|
||||
logged_artifact = self.experiment.get_artifact(artifact_path)
|
||||
artifact_save_dir = str(Path(self.opt.save_dir) / logged_artifact.name)
|
||||
logged_artifact.download(artifact_save_dir)
|
||||
|
||||
metadata = logged_artifact.metadata
|
||||
data_dict = metadata.copy()
|
||||
data_dict["path"] = artifact_save_dir
|
||||
|
||||
metadata_names = metadata.get("names")
|
||||
if isinstance(metadata_names, dict):
|
||||
data_dict["names"] = {int(k): v for k, v in metadata.get("names").items()}
|
||||
elif isinstance(metadata_names, list):
|
||||
data_dict["names"] = {int(k): v for k, v in zip(range(len(metadata_names)), metadata_names)}
|
||||
else:
|
||||
raise "Invalid 'names' field in dataset yaml file. Please use a list or dictionary"
|
||||
|
||||
return self.update_data_paths(data_dict)
|
||||
|
||||
def update_data_paths(self, data_dict):
|
||||
"""Updates data paths in the dataset dictionary, defaulting 'path' to an empty string if not present."""
|
||||
path = data_dict.get("path", "")
|
||||
|
||||
for split in ["train", "val", "test"]:
|
||||
if data_dict.get(split):
|
||||
split_path = data_dict.get(split)
|
||||
data_dict[split] = (
|
||||
f"{path}/{split_path}" if isinstance(split, str) else [f"{path}/{x}" for x in split_path]
|
||||
)
|
||||
|
||||
return data_dict
|
||||
|
||||
def on_pretrain_routine_end(self, paths):
|
||||
"""Called at the end of pretraining routine to handle paths if training is not being resumed."""
|
||||
if self.opt.resume:
|
||||
return
|
||||
|
||||
for path in paths:
|
||||
self.log_asset(str(path))
|
||||
|
||||
if self.upload_dataset and not self.resume:
|
||||
self.upload_dataset_artifact()
|
||||
|
||||
return
|
||||
|
||||
def on_train_start(self):
|
||||
"""Logs hyperparameters at the start of training."""
|
||||
self.log_parameters(self.hyp)
|
||||
|
||||
def on_train_epoch_start(self):
|
||||
"""Called at the start of each training epoch."""
|
||||
return
|
||||
|
||||
def on_train_epoch_end(self, epoch):
|
||||
"""Updates the current epoch in the experiment tracking at the end of each epoch."""
|
||||
self.experiment.curr_epoch = epoch
|
||||
|
||||
return
|
||||
|
||||
def on_train_batch_start(self):
|
||||
"""Called at the start of each training batch."""
|
||||
return
|
||||
|
||||
def on_train_batch_end(self, log_dict, step):
|
||||
"""Callback function that updates and logs metrics at the end of each training batch if conditions are met."""
|
||||
self.experiment.curr_step = step
|
||||
if self.log_batch_metrics and (step % self.comet_log_batch_interval == 0):
|
||||
self.log_metrics(log_dict, step=step)
|
||||
|
||||
return
|
||||
|
||||
def on_train_end(self, files, save_dir, last, best, epoch, results):
|
||||
"""Logs metadata and optionally saves model files at the end of training."""
|
||||
if self.comet_log_predictions:
|
||||
curr_epoch = self.experiment.curr_epoch
|
||||
self.experiment.log_asset_data(self.metadata_dict, "image-metadata.json", epoch=curr_epoch)
|
||||
|
||||
for f in files:
|
||||
self.log_asset(f, metadata={"epoch": epoch})
|
||||
self.log_asset(f"{save_dir}/results.csv", metadata={"epoch": epoch})
|
||||
|
||||
if not self.opt.evolve:
|
||||
model_path = str(best if best.exists() else last)
|
||||
name = Path(model_path).name
|
||||
if self.save_model:
|
||||
self.experiment.log_model(
|
||||
self.model_name,
|
||||
file_or_folder=model_path,
|
||||
file_name=name,
|
||||
overwrite=True,
|
||||
)
|
||||
|
||||
# Check if running Experiment with Comet Optimizer
|
||||
if hasattr(self.opt, "comet_optimizer_id"):
|
||||
metric = results.get(self.opt.comet_optimizer_metric)
|
||||
self.experiment.log_other("optimizer_metric_value", metric)
|
||||
|
||||
self.finish_run()
|
||||
|
||||
def on_val_start(self):
|
||||
"""Called at the start of validation, currently a placeholder with no functionality."""
|
||||
return
|
||||
|
||||
def on_val_batch_start(self):
|
||||
"""Placeholder called at the start of a validation batch with no current functionality."""
|
||||
return
|
||||
|
||||
def on_val_batch_end(self, batch_i, images, targets, paths, shapes, outputs):
|
||||
"""Callback executed at the end of a validation batch, conditionally logs predictions to Comet ML."""
|
||||
if not (self.comet_log_predictions and ((batch_i + 1) % self.comet_log_prediction_interval == 0)):
|
||||
return
|
||||
|
||||
for si, pred in enumerate(outputs):
|
||||
if len(pred) == 0:
|
||||
continue
|
||||
|
||||
image = images[si]
|
||||
labels = targets[targets[:, 0] == si, 1:]
|
||||
shape = shapes[si]
|
||||
path = paths[si]
|
||||
predn, labelsn = self.preprocess_prediction(image, labels, shape, pred)
|
||||
if labelsn is not None:
|
||||
self.log_predictions(image, labelsn, path, shape, predn)
|
||||
|
||||
return
|
||||
|
||||
def on_val_end(self, nt, tp, fp, p, r, f1, ap, ap50, ap_class, confusion_matrix):
|
||||
"""Logs per-class metrics to Comet.ml after validation if enabled and more than one class exists."""
|
||||
if self.comet_log_per_class_metrics and self.num_classes > 1:
|
||||
for i, c in enumerate(ap_class):
|
||||
class_name = self.class_names[c]
|
||||
self.experiment.log_metrics(
|
||||
{
|
||||
"mAP@.5": ap50[i],
|
||||
"mAP@.5:.95": ap[i],
|
||||
"precision": p[i],
|
||||
"recall": r[i],
|
||||
"f1": f1[i],
|
||||
"true_positives": tp[i],
|
||||
"false_positives": fp[i],
|
||||
"support": nt[c],
|
||||
},
|
||||
prefix=class_name,
|
||||
)
|
||||
|
||||
if self.comet_log_confusion_matrix:
|
||||
epoch = self.experiment.curr_epoch
|
||||
class_names = list(self.class_names.values())
|
||||
class_names.append("background")
|
||||
num_classes = len(class_names)
|
||||
|
||||
self.experiment.log_confusion_matrix(
|
||||
matrix=confusion_matrix.matrix,
|
||||
max_categories=num_classes,
|
||||
labels=class_names,
|
||||
epoch=epoch,
|
||||
column_label="Actual Category",
|
||||
row_label="Predicted Category",
|
||||
file_name=f"confusion-matrix-epoch-{epoch}.json",
|
||||
)
|
||||
|
||||
def on_fit_epoch_end(self, result, epoch):
|
||||
"""Logs metrics at the end of each training epoch."""
|
||||
self.log_metrics(result, epoch=epoch)
|
||||
|
||||
def on_model_save(self, last, epoch, final_epoch, best_fitness, fi):
|
||||
"""Callback to save model checkpoints periodically if conditions are met."""
|
||||
if ((epoch + 1) % self.opt.save_period == 0 and not final_epoch) and self.opt.save_period != -1:
|
||||
self.log_model(last.parent, self.opt, epoch, fi, best_model=best_fitness == fi)
|
||||
|
||||
def on_params_update(self, params):
|
||||
"""Logs updated parameters during training."""
|
||||
self.log_parameters(params)
|
||||
|
||||
def finish_run(self):
|
||||
"""Ends the current experiment and logs its completion."""
|
||||
self.experiment.end()
|
||||
@@ -1,151 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
import logging
|
||||
import os
|
||||
from urllib.parse import urlparse
|
||||
|
||||
try:
|
||||
import comet_ml
|
||||
except ImportError:
|
||||
comet_ml = None
|
||||
|
||||
import yaml
|
||||
|
||||
logger = logging.getLogger(__name__)
|
||||
|
||||
COMET_PREFIX = "comet://"
|
||||
COMET_MODEL_NAME = os.getenv("COMET_MODEL_NAME", "yolov5")
|
||||
COMET_DEFAULT_CHECKPOINT_FILENAME = os.getenv("COMET_DEFAULT_CHECKPOINT_FILENAME", "last.pt")
|
||||
|
||||
|
||||
def download_model_checkpoint(opt, experiment):
|
||||
"""Downloads YOLOv5 model checkpoint from Comet ML experiment, updating `opt.weights` with download path."""
|
||||
model_dir = f"{opt.project}/{experiment.name}"
|
||||
os.makedirs(model_dir, exist_ok=True)
|
||||
|
||||
model_name = COMET_MODEL_NAME
|
||||
model_asset_list = experiment.get_model_asset_list(model_name)
|
||||
|
||||
if len(model_asset_list) == 0:
|
||||
logger.error(f"COMET ERROR: No checkpoints found for model name : {model_name}")
|
||||
return
|
||||
|
||||
model_asset_list = sorted(
|
||||
model_asset_list,
|
||||
key=lambda x: x["step"],
|
||||
reverse=True,
|
||||
)
|
||||
logged_checkpoint_map = {asset["fileName"]: asset["assetId"] for asset in model_asset_list}
|
||||
|
||||
resource_url = urlparse(opt.weights)
|
||||
checkpoint_filename = resource_url.query
|
||||
|
||||
if checkpoint_filename:
|
||||
asset_id = logged_checkpoint_map.get(checkpoint_filename)
|
||||
else:
|
||||
asset_id = logged_checkpoint_map.get(COMET_DEFAULT_CHECKPOINT_FILENAME)
|
||||
checkpoint_filename = COMET_DEFAULT_CHECKPOINT_FILENAME
|
||||
|
||||
if asset_id is None:
|
||||
logger.error(f"COMET ERROR: Checkpoint {checkpoint_filename} not found in the given Experiment")
|
||||
return
|
||||
|
||||
try:
|
||||
logger.info(f"COMET INFO: Downloading checkpoint {checkpoint_filename}")
|
||||
asset_filename = checkpoint_filename
|
||||
|
||||
model_binary = experiment.get_asset(asset_id, return_type="binary", stream=False)
|
||||
model_download_path = f"{model_dir}/{asset_filename}"
|
||||
with open(model_download_path, "wb") as f:
|
||||
f.write(model_binary)
|
||||
|
||||
opt.weights = model_download_path
|
||||
|
||||
except Exception as e:
|
||||
logger.warning("COMET WARNING: Unable to download checkpoint from Comet")
|
||||
logger.exception(e)
|
||||
|
||||
|
||||
def set_opt_parameters(opt, experiment):
|
||||
"""
|
||||
Update the opts Namespace with parameters from Comet's ExistingExperiment when resuming a run.
|
||||
|
||||
Args:
|
||||
opt (argparse.Namespace): Namespace of command line options
|
||||
experiment (comet_ml.APIExperiment): Comet API Experiment object
|
||||
"""
|
||||
asset_list = experiment.get_asset_list()
|
||||
resume_string = opt.resume
|
||||
|
||||
for asset in asset_list:
|
||||
if asset["fileName"] == "opt.yaml":
|
||||
asset_id = asset["assetId"]
|
||||
asset_binary = experiment.get_asset(asset_id, return_type="binary", stream=False)
|
||||
opt_dict = yaml.safe_load(asset_binary)
|
||||
for key, value in opt_dict.items():
|
||||
setattr(opt, key, value)
|
||||
opt.resume = resume_string
|
||||
|
||||
# Save hyperparameters to YAML file
|
||||
# Necessary to pass checks in training script
|
||||
save_dir = f"{opt.project}/{experiment.name}"
|
||||
os.makedirs(save_dir, exist_ok=True)
|
||||
|
||||
hyp_yaml_path = f"{save_dir}/hyp.yaml"
|
||||
with open(hyp_yaml_path, "w") as f:
|
||||
yaml.dump(opt.hyp, f)
|
||||
opt.hyp = hyp_yaml_path
|
||||
|
||||
|
||||
def check_comet_weights(opt):
|
||||
"""
|
||||
Downloads model weights from Comet and updates the weights path to point to saved weights location.
|
||||
|
||||
Args:
|
||||
opt (argparse.Namespace): Command Line arguments passed
|
||||
to YOLOv5 training script
|
||||
|
||||
Returns:
|
||||
None/bool: Return True if weights are successfully downloaded
|
||||
else return None
|
||||
"""
|
||||
if comet_ml is None:
|
||||
return
|
||||
|
||||
if isinstance(opt.weights, str) and opt.weights.startswith(COMET_PREFIX):
|
||||
api = comet_ml.API()
|
||||
resource = urlparse(opt.weights)
|
||||
experiment_path = f"{resource.netloc}{resource.path}"
|
||||
experiment = api.get(experiment_path)
|
||||
download_model_checkpoint(opt, experiment)
|
||||
return True
|
||||
|
||||
return None
|
||||
|
||||
|
||||
def check_comet_resume(opt):
|
||||
"""
|
||||
Restores run parameters to its original state based on the model checkpoint and logged Experiment parameters.
|
||||
|
||||
Args:
|
||||
opt (argparse.Namespace): Command Line arguments passed
|
||||
to YOLOv5 training script
|
||||
|
||||
Returns:
|
||||
None/bool: Return True if the run is restored successfully
|
||||
else return None
|
||||
"""
|
||||
if comet_ml is None:
|
||||
return
|
||||
|
||||
if isinstance(opt.resume, str) and opt.resume.startswith(COMET_PREFIX):
|
||||
api = comet_ml.API()
|
||||
resource = urlparse(opt.resume)
|
||||
experiment_path = f"{resource.netloc}{resource.path}"
|
||||
experiment = api.get(experiment_path)
|
||||
set_opt_parameters(opt, experiment)
|
||||
download_model_checkpoint(opt, experiment)
|
||||
|
||||
return True
|
||||
|
||||
return None
|
||||
@@ -1,126 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
import argparse
|
||||
import json
|
||||
import logging
|
||||
import os
|
||||
import sys
|
||||
from pathlib import Path
|
||||
|
||||
import comet_ml
|
||||
|
||||
logger = logging.getLogger(__name__)
|
||||
|
||||
FILE = Path(__file__).resolve()
|
||||
ROOT = FILE.parents[3] # YOLOv5 root directory
|
||||
if str(ROOT) not in sys.path:
|
||||
sys.path.append(str(ROOT)) # add ROOT to PATH
|
||||
|
||||
from train import train
|
||||
from utils.callbacks import Callbacks
|
||||
from utils.general import increment_path
|
||||
from utils.torch_utils import select_device
|
||||
|
||||
# Project Configuration
|
||||
config = comet_ml.config.get_config()
|
||||
COMET_PROJECT_NAME = config.get_string(os.getenv("COMET_PROJECT_NAME"), "comet.project_name", default="yolov5")
|
||||
|
||||
|
||||
def get_args(known=False):
|
||||
"""Parses command-line arguments for YOLOv5 training, supporting configuration of weights, data paths,
|
||||
hyperparameters, and more.
|
||||
"""
|
||||
parser = argparse.ArgumentParser()
|
||||
parser.add_argument("--weights", type=str, default=ROOT / "yolov5s.pt", help="initial weights path")
|
||||
parser.add_argument("--cfg", type=str, default="", help="model.yaml path")
|
||||
parser.add_argument("--data", type=str, default=ROOT / "data/coco128.yaml", help="dataset.yaml path")
|
||||
parser.add_argument("--hyp", type=str, default=ROOT / "data/hyps/hyp.scratch-low.yaml", help="hyperparameters path")
|
||||
parser.add_argument("--epochs", type=int, default=300, help="total training epochs")
|
||||
parser.add_argument("--batch-size", type=int, default=16, help="total batch size for all GPUs, -1 for autobatch")
|
||||
parser.add_argument("--imgsz", "--img", "--img-size", type=int, default=640, help="train, val image size (pixels)")
|
||||
parser.add_argument("--rect", action="store_true", help="rectangular training")
|
||||
parser.add_argument("--resume", nargs="?", const=True, default=False, help="resume most recent training")
|
||||
parser.add_argument("--nosave", action="store_true", help="only save final checkpoint")
|
||||
parser.add_argument("--noval", action="store_true", help="only validate final epoch")
|
||||
parser.add_argument("--noautoanchor", action="store_true", help="disable AutoAnchor")
|
||||
parser.add_argument("--noplots", action="store_true", help="save no plot files")
|
||||
parser.add_argument("--evolve", type=int, nargs="?", const=300, help="evolve hyperparameters for x generations")
|
||||
parser.add_argument("--bucket", type=str, default="", help="gsutil bucket")
|
||||
parser.add_argument("--cache", type=str, nargs="?", const="ram", help='--cache images in "ram" (default) or "disk"')
|
||||
parser.add_argument("--image-weights", action="store_true", help="use weighted image selection for training")
|
||||
parser.add_argument("--device", default="", help="cuda device, i.e. 0 or 0,1,2,3 or cpu")
|
||||
parser.add_argument("--multi-scale", action="store_true", help="vary img-size +/- 50%%")
|
||||
parser.add_argument("--single-cls", action="store_true", help="train multi-class data as single-class")
|
||||
parser.add_argument("--optimizer", type=str, choices=["SGD", "Adam", "AdamW"], default="SGD", help="optimizer")
|
||||
parser.add_argument("--sync-bn", action="store_true", help="use SyncBatchNorm, only available in DDP mode")
|
||||
parser.add_argument("--workers", type=int, default=8, help="max dataloader workers (per RANK in DDP mode)")
|
||||
parser.add_argument("--project", default=ROOT / "runs/train", help="save to project/name")
|
||||
parser.add_argument("--name", default="exp", help="save to project/name")
|
||||
parser.add_argument("--exist-ok", action="store_true", help="existing project/name ok, do not increment")
|
||||
parser.add_argument("--quad", action="store_true", help="quad dataloader")
|
||||
parser.add_argument("--cos-lr", action="store_true", help="cosine LR scheduler")
|
||||
parser.add_argument("--label-smoothing", type=float, default=0.0, help="Label smoothing epsilon")
|
||||
parser.add_argument("--patience", type=int, default=100, help="EarlyStopping patience (epochs without improvement)")
|
||||
parser.add_argument("--freeze", nargs="+", type=int, default=[0], help="Freeze layers: backbone=10, first3=0 1 2")
|
||||
parser.add_argument("--save-period", type=int, default=-1, help="Save checkpoint every x epochs (disabled if < 1)")
|
||||
parser.add_argument("--seed", type=int, default=0, help="Global training seed")
|
||||
parser.add_argument("--local_rank", type=int, default=-1, help="Automatic DDP Multi-GPU argument, do not modify")
|
||||
|
||||
# Weights & Biases arguments
|
||||
parser.add_argument("--entity", default=None, help="W&B: Entity")
|
||||
parser.add_argument("--upload_dataset", nargs="?", const=True, default=False, help='W&B: Upload data, "val" option')
|
||||
parser.add_argument("--bbox_interval", type=int, default=-1, help="W&B: Set bounding-box image logging interval")
|
||||
parser.add_argument("--artifact_alias", type=str, default="latest", help="W&B: Version of dataset artifact to use")
|
||||
|
||||
# Comet Arguments
|
||||
parser.add_argument("--comet_optimizer_config", type=str, help="Comet: Path to a Comet Optimizer Config File.")
|
||||
parser.add_argument("--comet_optimizer_id", type=str, help="Comet: ID of the Comet Optimizer sweep.")
|
||||
parser.add_argument("--comet_optimizer_objective", type=str, help="Comet: Set to 'minimize' or 'maximize'.")
|
||||
parser.add_argument("--comet_optimizer_metric", type=str, help="Comet: Metric to Optimize.")
|
||||
parser.add_argument(
|
||||
"--comet_optimizer_workers",
|
||||
type=int,
|
||||
default=1,
|
||||
help="Comet: Number of Parallel Workers to use with the Comet Optimizer.",
|
||||
)
|
||||
|
||||
return parser.parse_known_args()[0] if known else parser.parse_args()
|
||||
|
||||
|
||||
def run(parameters, opt):
|
||||
"""Executes YOLOv5 training with given hyperparameters and options, setting up device and training directories."""
|
||||
hyp_dict = {k: v for k, v in parameters.items() if k not in ["epochs", "batch_size"]}
|
||||
|
||||
opt.save_dir = str(increment_path(Path(opt.project) / opt.name, exist_ok=opt.exist_ok or opt.evolve))
|
||||
opt.batch_size = parameters.get("batch_size")
|
||||
opt.epochs = parameters.get("epochs")
|
||||
|
||||
device = select_device(opt.device, batch_size=opt.batch_size)
|
||||
train(hyp_dict, opt, device, callbacks=Callbacks())
|
||||
|
||||
|
||||
if __name__ == "__main__":
|
||||
opt = get_args(known=True)
|
||||
|
||||
opt.weights = str(opt.weights)
|
||||
opt.cfg = str(opt.cfg)
|
||||
opt.data = str(opt.data)
|
||||
opt.project = str(opt.project)
|
||||
|
||||
optimizer_id = os.getenv("COMET_OPTIMIZER_ID")
|
||||
if optimizer_id is None:
|
||||
with open(opt.comet_optimizer_config) as f:
|
||||
optimizer_config = json.load(f)
|
||||
optimizer = comet_ml.Optimizer(optimizer_config)
|
||||
else:
|
||||
optimizer = comet_ml.Optimizer(optimizer_id)
|
||||
|
||||
opt.comet_optimizer_id = optimizer.id
|
||||
status = optimizer.status()
|
||||
|
||||
opt.comet_optimizer_objective = status["spec"]["objective"]
|
||||
opt.comet_optimizer_metric = status["spec"]["metric"]
|
||||
|
||||
logger.info("COMET INFO: Starting Hyperparameter Sweep")
|
||||
for parameter in optimizer.get_parameters():
|
||||
run(parameter["parameters"], opt)
|
||||
@@ -1,135 +0,0 @@
|
||||
{
|
||||
"algorithm": "random",
|
||||
"parameters": {
|
||||
"anchor_t": {
|
||||
"type": "discrete",
|
||||
"values": [2, 8]
|
||||
},
|
||||
"batch_size": {
|
||||
"type": "discrete",
|
||||
"values": [16, 32, 64]
|
||||
},
|
||||
"box": {
|
||||
"type": "discrete",
|
||||
"values": [0.02, 0.2]
|
||||
},
|
||||
"cls": {
|
||||
"type": "discrete",
|
||||
"values": [0.2]
|
||||
},
|
||||
"cls_pw": {
|
||||
"type": "discrete",
|
||||
"values": [0.5]
|
||||
},
|
||||
"copy_paste": {
|
||||
"type": "discrete",
|
||||
"values": [1]
|
||||
},
|
||||
"degrees": {
|
||||
"type": "discrete",
|
||||
"values": [0, 45]
|
||||
},
|
||||
"epochs": {
|
||||
"type": "discrete",
|
||||
"values": [5]
|
||||
},
|
||||
"fl_gamma": {
|
||||
"type": "discrete",
|
||||
"values": [0]
|
||||
},
|
||||
"fliplr": {
|
||||
"type": "discrete",
|
||||
"values": [0]
|
||||
},
|
||||
"flipud": {
|
||||
"type": "discrete",
|
||||
"values": [0]
|
||||
},
|
||||
"hsv_h": {
|
||||
"type": "discrete",
|
||||
"values": [0]
|
||||
},
|
||||
"hsv_s": {
|
||||
"type": "discrete",
|
||||
"values": [0]
|
||||
},
|
||||
"hsv_v": {
|
||||
"type": "discrete",
|
||||
"values": [0]
|
||||
},
|
||||
"iou_t": {
|
||||
"type": "discrete",
|
||||
"values": [0.7]
|
||||
},
|
||||
"lr0": {
|
||||
"type": "discrete",
|
||||
"values": [1e-5, 0.1]
|
||||
},
|
||||
"lrf": {
|
||||
"type": "discrete",
|
||||
"values": [0.01, 1]
|
||||
},
|
||||
"mixup": {
|
||||
"type": "discrete",
|
||||
"values": [1]
|
||||
},
|
||||
"momentum": {
|
||||
"type": "discrete",
|
||||
"values": [0.6]
|
||||
},
|
||||
"mosaic": {
|
||||
"type": "discrete",
|
||||
"values": [0]
|
||||
},
|
||||
"obj": {
|
||||
"type": "discrete",
|
||||
"values": [0.2]
|
||||
},
|
||||
"obj_pw": {
|
||||
"type": "discrete",
|
||||
"values": [0.5]
|
||||
},
|
||||
"optimizer": {
|
||||
"type": "categorical",
|
||||
"values": ["SGD", "Adam", "AdamW"]
|
||||
},
|
||||
"perspective": {
|
||||
"type": "discrete",
|
||||
"values": [0]
|
||||
},
|
||||
"scale": {
|
||||
"type": "discrete",
|
||||
"values": [0]
|
||||
},
|
||||
"shear": {
|
||||
"type": "discrete",
|
||||
"values": [0]
|
||||
},
|
||||
"translate": {
|
||||
"type": "discrete",
|
||||
"values": [0]
|
||||
},
|
||||
"warmup_bias_lr": {
|
||||
"type": "discrete",
|
||||
"values": [0, 0.2]
|
||||
},
|
||||
"warmup_epochs": {
|
||||
"type": "discrete",
|
||||
"values": [5]
|
||||
},
|
||||
"warmup_momentum": {
|
||||
"type": "discrete",
|
||||
"values": [0, 0.95]
|
||||
},
|
||||
"weight_decay": {
|
||||
"type": "discrete",
|
||||
"values": [0, 0.001]
|
||||
}
|
||||
},
|
||||
"spec": {
|
||||
"maxCombo": 0,
|
||||
"metric": "metrics/mAP_0.5",
|
||||
"objective": "maximize"
|
||||
},
|
||||
"trials": 1
|
||||
}
|
||||
@@ -1 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
@@ -1,210 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# WARNING ⚠️ wandb is deprecated and will be removed in future release.
|
||||
# See supported integrations at https://github.com/ultralytics/yolov5#integrations
|
||||
|
||||
import logging
|
||||
import os
|
||||
import sys
|
||||
from contextlib import contextmanager
|
||||
from pathlib import Path
|
||||
|
||||
from utils.general import LOGGER, colorstr
|
||||
|
||||
FILE = Path(__file__).resolve()
|
||||
ROOT = FILE.parents[3] # YOLOv5 root directory
|
||||
if str(ROOT) not in sys.path:
|
||||
sys.path.append(str(ROOT)) # add ROOT to PATH
|
||||
RANK = int(os.getenv("RANK", -1))
|
||||
DEPRECATION_WARNING = (
|
||||
f"{colorstr('wandb')}: WARNING ⚠️ wandb is deprecated and will be removed in a future release. "
|
||||
f"See supported integrations at https://github.com/ultralytics/yolov5#integrations."
|
||||
)
|
||||
|
||||
try:
|
||||
import wandb
|
||||
|
||||
assert hasattr(wandb, "__version__") # verify package import not local dir
|
||||
LOGGER.warning(DEPRECATION_WARNING)
|
||||
except (ImportError, AssertionError):
|
||||
wandb = None
|
||||
|
||||
|
||||
class WandbLogger:
|
||||
"""
|
||||
Log training runs, datasets, models, and predictions to Weights & Biases.
|
||||
|
||||
This logger sends information to W&B at wandb.ai. By default, this information includes hyperparameters, system
|
||||
configuration and metrics, model metrics, and basic data metrics and analyses.
|
||||
|
||||
By providing additional command line arguments to train.py, datasets, models and predictions can also be logged.
|
||||
|
||||
For more on how this logger is used, see the Weights & Biases documentation:
|
||||
https://docs.wandb.com/guides/integrations/yolov5
|
||||
"""
|
||||
|
||||
def __init__(self, opt, run_id=None, job_type="Training"):
|
||||
"""
|
||||
- Initialize WandbLogger instance
|
||||
- Upload dataset if opt.upload_dataset is True
|
||||
- Setup training processes if job_type is 'Training'.
|
||||
|
||||
Arguments:
|
||||
opt (namespace) -- Commandline arguments for this run
|
||||
run_id (str) -- Run ID of W&B run to be resumed
|
||||
job_type (str) -- To set the job_type for this run
|
||||
|
||||
"""
|
||||
# Pre-training routine --
|
||||
self.job_type = job_type
|
||||
self.wandb, self.wandb_run = wandb, wandb.run if wandb else None
|
||||
self.val_artifact, self.train_artifact = None, None
|
||||
self.train_artifact_path, self.val_artifact_path = None, None
|
||||
self.result_artifact = None
|
||||
self.val_table, self.result_table = None, None
|
||||
self.max_imgs_to_log = 16
|
||||
self.data_dict = None
|
||||
if self.wandb:
|
||||
self.wandb_run = wandb.run or wandb.init(
|
||||
config=opt,
|
||||
resume="allow",
|
||||
project="YOLOv5" if opt.project == "runs/train" else Path(opt.project).stem,
|
||||
entity=opt.entity,
|
||||
name=opt.name if opt.name != "exp" else None,
|
||||
job_type=job_type,
|
||||
id=run_id,
|
||||
allow_val_change=True,
|
||||
)
|
||||
|
||||
if self.wandb_run and self.job_type == "Training":
|
||||
if isinstance(opt.data, dict):
|
||||
# This means another dataset manager has already processed the dataset info (e.g. ClearML)
|
||||
# and they will have stored the already processed dict in opt.data
|
||||
self.data_dict = opt.data
|
||||
self.setup_training(opt)
|
||||
|
||||
def setup_training(self, opt):
|
||||
"""
|
||||
Setup the necessary processes for training YOLO models:
|
||||
- Attempt to download model checkpoint and dataset artifacts if opt.resume stats with WANDB_ARTIFACT_PREFIX
|
||||
- Update data_dict, to contain info of previous run if resumed and the paths of dataset artifact if downloaded
|
||||
- Setup log_dict, initialize bbox_interval.
|
||||
|
||||
Arguments:
|
||||
opt (namespace) -- commandline arguments for this run
|
||||
|
||||
"""
|
||||
self.log_dict, self.current_epoch = {}, 0
|
||||
self.bbox_interval = opt.bbox_interval
|
||||
if isinstance(opt.resume, str):
|
||||
model_dir, _ = self.download_model_artifact(opt)
|
||||
if model_dir:
|
||||
self.weights = Path(model_dir) / "last.pt"
|
||||
config = self.wandb_run.config
|
||||
opt.weights, opt.save_period, opt.batch_size, opt.bbox_interval, opt.epochs, opt.hyp, opt.imgsz = (
|
||||
str(self.weights),
|
||||
config.save_period,
|
||||
config.batch_size,
|
||||
config.bbox_interval,
|
||||
config.epochs,
|
||||
config.hyp,
|
||||
config.imgsz,
|
||||
)
|
||||
|
||||
if opt.bbox_interval == -1:
|
||||
self.bbox_interval = opt.bbox_interval = (opt.epochs // 10) if opt.epochs > 10 else 1
|
||||
if opt.evolve or opt.noplots:
|
||||
self.bbox_interval = opt.bbox_interval = opt.epochs + 1 # disable bbox_interval
|
||||
|
||||
def log_model(self, path, opt, epoch, fitness_score, best_model=False):
|
||||
"""
|
||||
Log the model checkpoint as W&B artifact.
|
||||
|
||||
Arguments:
|
||||
path (Path) -- Path of directory containing the checkpoints
|
||||
opt (namespace) -- Command line arguments for this run
|
||||
epoch (int) -- Current epoch number
|
||||
fitness_score (float) -- fitness score for current epoch
|
||||
best_model (boolean) -- Boolean representing if the current checkpoint is the best yet.
|
||||
"""
|
||||
model_artifact = wandb.Artifact(
|
||||
f"run_{wandb.run.id}_model",
|
||||
type="model",
|
||||
metadata={
|
||||
"original_url": str(path),
|
||||
"epochs_trained": epoch + 1,
|
||||
"save period": opt.save_period,
|
||||
"project": opt.project,
|
||||
"total_epochs": opt.epochs,
|
||||
"fitness_score": fitness_score,
|
||||
},
|
||||
)
|
||||
model_artifact.add_file(str(path / "last.pt"), name="last.pt")
|
||||
wandb.log_artifact(
|
||||
model_artifact,
|
||||
aliases=[
|
||||
"latest",
|
||||
"last",
|
||||
f"epoch {str(self.current_epoch)}",
|
||||
"best" if best_model else "",
|
||||
],
|
||||
)
|
||||
LOGGER.info(f"Saving model artifact on epoch {epoch + 1}")
|
||||
|
||||
def val_one_image(self, pred, predn, path, names, im):
|
||||
"""Evaluates model prediction for a single image, returning metrics and visualizations."""
|
||||
pass
|
||||
|
||||
def log(self, log_dict):
|
||||
"""
|
||||
Save the metrics to the logging dictionary.
|
||||
|
||||
Arguments:
|
||||
log_dict (Dict) -- metrics/media to be logged in current step
|
||||
"""
|
||||
if self.wandb_run:
|
||||
for key, value in log_dict.items():
|
||||
self.log_dict[key] = value
|
||||
|
||||
def end_epoch(self):
|
||||
"""
|
||||
Commit the log_dict, model artifacts and Tables to W&B and flush the log_dict.
|
||||
|
||||
Arguments:
|
||||
best_result (boolean): Boolean representing if the result of this evaluation is best or not
|
||||
"""
|
||||
if self.wandb_run:
|
||||
with all_logging_disabled():
|
||||
try:
|
||||
wandb.log(self.log_dict)
|
||||
except BaseException as e:
|
||||
LOGGER.info(
|
||||
f"An error occurred in wandb logger. The training will proceed without interruption. More info\n{e}"
|
||||
)
|
||||
self.wandb_run.finish()
|
||||
self.wandb_run = None
|
||||
self.log_dict = {}
|
||||
|
||||
def finish_run(self):
|
||||
"""Log metrics if any and finish the current W&B run."""
|
||||
if self.wandb_run:
|
||||
if self.log_dict:
|
||||
with all_logging_disabled():
|
||||
wandb.log(self.log_dict)
|
||||
wandb.run.finish()
|
||||
LOGGER.warning(DEPRECATION_WARNING)
|
||||
|
||||
|
||||
@contextmanager
|
||||
def all_logging_disabled(highest_level=logging.CRITICAL):
|
||||
"""Source - https://gist.github.com/simon-weber/7853144
|
||||
A context manager that will prevent any logging messages triggered during the body from being processed.
|
||||
:param highest_level: the maximum logging level in use.
|
||||
This would only need to be changed if a custom level greater than CRITICAL is defined.
|
||||
"""
|
||||
previous_level = logging.root.manager.disable
|
||||
logging.disable(highest_level)
|
||||
try:
|
||||
yield
|
||||
finally:
|
||||
logging.disable(previous_level)
|
||||
@@ -1,254 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Loss functions."""
|
||||
|
||||
import torch
|
||||
import torch.nn as nn
|
||||
|
||||
from utils.yolov5.utils.metrics import bbox_iou
|
||||
from utils.yolov5.utils.torch_utils import de_parallel
|
||||
|
||||
|
||||
def smooth_BCE(eps=0.1):
|
||||
"""Returns label smoothing BCE targets for reducing overfitting; pos: `1.0 - 0.5*eps`, neg: `0.5*eps`. For details see https://github.com/ultralytics/yolov3/issues/238#issuecomment-598028441."""
|
||||
return 1.0 - 0.5 * eps, 0.5 * eps
|
||||
|
||||
|
||||
class BCEBlurWithLogitsLoss(nn.Module):
|
||||
"""Modified BCEWithLogitsLoss to reduce missing label effects in YOLOv5 training with optional alpha smoothing."""
|
||||
|
||||
def __init__(self, alpha=0.05):
|
||||
"""Initializes a modified BCEWithLogitsLoss with reduced missing label effects, taking optional alpha smoothing
|
||||
parameter.
|
||||
"""
|
||||
super().__init__()
|
||||
self.loss_fcn = nn.BCEWithLogitsLoss(reduction="none") # must be nn.BCEWithLogitsLoss()
|
||||
self.alpha = alpha
|
||||
|
||||
def forward(self, pred, true):
|
||||
"""Computes modified BCE loss for YOLOv5 with reduced missing label effects, taking pred and true tensors,
|
||||
returns mean loss.
|
||||
"""
|
||||
loss = self.loss_fcn(pred, true)
|
||||
pred = torch.sigmoid(pred) # prob from logits
|
||||
dx = pred - true # reduce only missing label effects
|
||||
# dx = (pred - true).abs() # reduce missing label and false label effects
|
||||
alpha_factor = 1 - torch.exp((dx - 1) / (self.alpha + 1e-4))
|
||||
loss *= alpha_factor
|
||||
return loss.mean()
|
||||
|
||||
|
||||
class FocalLoss(nn.Module):
|
||||
"""Applies focal loss to address class imbalance by modifying BCEWithLogitsLoss with gamma and alpha parameters."""
|
||||
|
||||
def __init__(self, loss_fcn, gamma=1.5, alpha=0.25):
|
||||
"""Initializes FocalLoss with specified loss function, gamma, and alpha values; modifies loss reduction to
|
||||
'none'.
|
||||
"""
|
||||
super().__init__()
|
||||
self.loss_fcn = loss_fcn # must be nn.BCEWithLogitsLoss()
|
||||
self.gamma = gamma
|
||||
self.alpha = alpha
|
||||
self.reduction = loss_fcn.reduction
|
||||
self.loss_fcn.reduction = "none" # required to apply FL to each element
|
||||
|
||||
def forward(self, pred, true):
|
||||
"""Calculates the focal loss between predicted and true labels using a modified BCEWithLogitsLoss."""
|
||||
loss = self.loss_fcn(pred, true)
|
||||
# p_t = torch.exp(-loss)
|
||||
# loss *= self.alpha * (1.000001 - p_t) ** self.gamma # non-zero power for gradient stability
|
||||
|
||||
# TF implementation https://github.com/tensorflow/addons/blob/v0.7.1/tensorflow_addons/losses/focal_loss.py
|
||||
pred_prob = torch.sigmoid(pred) # prob from logits
|
||||
p_t = true * pred_prob + (1 - true) * (1 - pred_prob)
|
||||
alpha_factor = true * self.alpha + (1 - true) * (1 - self.alpha)
|
||||
modulating_factor = (1.0 - p_t) ** self.gamma
|
||||
loss *= alpha_factor * modulating_factor
|
||||
|
||||
if self.reduction == "mean":
|
||||
return loss.mean()
|
||||
elif self.reduction == "sum":
|
||||
return loss.sum()
|
||||
else: # 'none'
|
||||
return loss
|
||||
|
||||
|
||||
class QFocalLoss(nn.Module):
|
||||
"""Implements Quality Focal Loss to address class imbalance by modulating loss based on prediction confidence."""
|
||||
|
||||
def __init__(self, loss_fcn, gamma=1.5, alpha=0.25):
|
||||
"""Initializes Quality Focal Loss with given loss function, gamma, alpha; modifies reduction to 'none'."""
|
||||
super().__init__()
|
||||
self.loss_fcn = loss_fcn # must be nn.BCEWithLogitsLoss()
|
||||
self.gamma = gamma
|
||||
self.alpha = alpha
|
||||
self.reduction = loss_fcn.reduction
|
||||
self.loss_fcn.reduction = "none" # required to apply FL to each element
|
||||
|
||||
def forward(self, pred, true):
|
||||
"""Computes the focal loss between `pred` and `true` using BCEWithLogitsLoss, adjusting for imbalance with
|
||||
`gamma` and `alpha`.
|
||||
"""
|
||||
loss = self.loss_fcn(pred, true)
|
||||
|
||||
pred_prob = torch.sigmoid(pred) # prob from logits
|
||||
alpha_factor = true * self.alpha + (1 - true) * (1 - self.alpha)
|
||||
modulating_factor = torch.abs(true - pred_prob) ** self.gamma
|
||||
loss *= alpha_factor * modulating_factor
|
||||
|
||||
if self.reduction == "mean":
|
||||
return loss.mean()
|
||||
elif self.reduction == "sum":
|
||||
return loss.sum()
|
||||
else: # 'none'
|
||||
return loss
|
||||
|
||||
|
||||
class ComputeLoss:
|
||||
"""Computes the total loss for YOLOv5 model predictions, including classification, box, and objectness losses."""
|
||||
|
||||
sort_obj_iou = False
|
||||
|
||||
# Compute losses
|
||||
def __init__(self, model, autobalance=False):
|
||||
"""Initializes ComputeLoss with model and autobalance option, autobalances losses if True."""
|
||||
device = next(model.parameters()).device # get model device
|
||||
h = model.hyp # hyperparameters
|
||||
|
||||
# Define criteria
|
||||
BCEcls = nn.BCEWithLogitsLoss(pos_weight=torch.tensor([h["cls_pw"]], device=device))
|
||||
BCEobj = nn.BCEWithLogitsLoss(pos_weight=torch.tensor([h["obj_pw"]], device=device))
|
||||
|
||||
# Class label smoothing https://arxiv.org/pdf/1902.04103.pdf eqn 3
|
||||
self.cp, self.cn = smooth_BCE(eps=h.get("label_smoothing", 0.0)) # positive, negative BCE targets
|
||||
|
||||
# Focal loss
|
||||
g = h["fl_gamma"] # focal loss gamma
|
||||
if g > 0:
|
||||
BCEcls, BCEobj = FocalLoss(BCEcls, g), FocalLoss(BCEobj, g)
|
||||
|
||||
m = de_parallel(model).model[-1] # Detect() module
|
||||
self.balance = {3: [4.0, 1.0, 0.4]}.get(m.nl, [4.0, 1.0, 0.25, 0.06, 0.02]) # P3-P7
|
||||
self.ssi = list(m.stride).index(16) if autobalance else 0 # stride 16 index
|
||||
self.BCEcls, self.BCEobj, self.gr, self.hyp, self.autobalance = BCEcls, BCEobj, 1.0, h, autobalance
|
||||
self.na = m.na # number of anchors
|
||||
self.nc = m.nc # number of classes
|
||||
self.nl = m.nl # number of layers
|
||||
self.anchors = m.anchors
|
||||
self.device = device
|
||||
|
||||
def __call__(self, p, targets): # predictions, targets
|
||||
"""Performs forward pass, calculating class, box, and object loss for given predictions and targets."""
|
||||
lcls = torch.zeros(1, device=self.device) # class loss
|
||||
lbox = torch.zeros(1, device=self.device) # box loss
|
||||
lobj = torch.zeros(1, device=self.device) # object loss
|
||||
tcls, tbox, indices, anchors = self.build_targets(p, targets) # targets
|
||||
|
||||
# Losses
|
||||
for i, pi in enumerate(p): # layer index, layer predictions
|
||||
b, a, gj, gi = indices[i] # image, anchor, gridy, gridx
|
||||
tobj = torch.zeros(pi.shape[:4], dtype=pi.dtype, device=self.device) # target obj
|
||||
|
||||
if n := b.shape[0]:
|
||||
# pxy, pwh, _, pcls = pi[b, a, gj, gi].tensor_split((2, 4, 5), dim=1) # faster, requires torch 1.8.0
|
||||
pxy, pwh, _, pcls = pi[b, a, gj, gi].split((2, 2, 1, self.nc), 1) # target-subset of predictions
|
||||
|
||||
# Regression
|
||||
pxy = pxy.sigmoid() * 2 - 0.5
|
||||
pwh = (pwh.sigmoid() * 2) ** 2 * anchors[i]
|
||||
pbox = torch.cat((pxy, pwh), 1) # predicted box
|
||||
iou = bbox_iou(pbox, tbox[i], CIoU=True).squeeze() # iou(prediction, target)
|
||||
lbox += (1.0 - iou).mean() # iou loss
|
||||
|
||||
# Objectness
|
||||
iou = iou.detach().clamp(0).type(tobj.dtype)
|
||||
if self.sort_obj_iou:
|
||||
j = iou.argsort()
|
||||
b, a, gj, gi, iou = b[j], a[j], gj[j], gi[j], iou[j]
|
||||
if self.gr < 1:
|
||||
iou = (1.0 - self.gr) + self.gr * iou
|
||||
tobj[b, a, gj, gi] = iou # iou ratio
|
||||
|
||||
# Classification
|
||||
if self.nc > 1: # cls loss (only if multiple classes)
|
||||
t = torch.full_like(pcls, self.cn, device=self.device) # targets
|
||||
t[range(n), tcls[i]] = self.cp
|
||||
lcls += self.BCEcls(pcls, t) # BCE
|
||||
|
||||
obji = self.BCEobj(pi[..., 4], tobj)
|
||||
lobj += obji * self.balance[i] # obj loss
|
||||
if self.autobalance:
|
||||
self.balance[i] = self.balance[i] * 0.9999 + 0.0001 / obji.detach().item()
|
||||
|
||||
if self.autobalance:
|
||||
self.balance = [x / self.balance[self.ssi] for x in self.balance]
|
||||
lbox *= self.hyp["box"]
|
||||
lobj *= self.hyp["obj"]
|
||||
lcls *= self.hyp["cls"]
|
||||
bs = tobj.shape[0] # batch size
|
||||
|
||||
return (lbox + lobj + lcls) * bs, torch.cat((lbox, lobj, lcls)).detach()
|
||||
|
||||
def build_targets(self, p, targets):
|
||||
"""Prepares model targets from input targets (image,class,x,y,w,h) for loss computation, returning class, box,
|
||||
indices, and anchors.
|
||||
"""
|
||||
na, nt = self.na, targets.shape[0] # number of anchors, targets
|
||||
tcls, tbox, indices, anch = [], [], [], []
|
||||
gain = torch.ones(7, device=self.device) # normalized to gridspace gain
|
||||
ai = torch.arange(na, device=self.device).float().view(na, 1).repeat(1, nt) # same as .repeat_interleave(nt)
|
||||
targets = torch.cat((targets.repeat(na, 1, 1), ai[..., None]), 2) # append anchor indices
|
||||
|
||||
g = 0.5 # bias
|
||||
off = (
|
||||
torch.tensor(
|
||||
[
|
||||
[0, 0],
|
||||
[1, 0],
|
||||
[0, 1],
|
||||
[-1, 0],
|
||||
[0, -1], # j,k,l,m
|
||||
# [1, 1], [1, -1], [-1, 1], [-1, -1], # jk,jm,lk,lm
|
||||
],
|
||||
device=self.device,
|
||||
).float()
|
||||
* g
|
||||
) # offsets
|
||||
|
||||
for i in range(self.nl):
|
||||
anchors, shape = self.anchors[i], p[i].shape
|
||||
gain[2:6] = torch.tensor(shape)[[3, 2, 3, 2]] # xyxy gain
|
||||
|
||||
# Match targets to anchors
|
||||
t = targets * gain # shape(3,n,7)
|
||||
if nt:
|
||||
# Matches
|
||||
r = t[..., 4:6] / anchors[:, None] # wh ratio
|
||||
j = torch.max(r, 1 / r).max(2)[0] < self.hyp["anchor_t"] # compare
|
||||
# j = wh_iou(anchors, t[:, 4:6]) > model.hyp['iou_t'] # iou(3,n)=wh_iou(anchors(3,2), gwh(n,2))
|
||||
t = t[j] # filter
|
||||
|
||||
# Offsets
|
||||
gxy = t[:, 2:4] # grid xy
|
||||
gxi = gain[[2, 3]] - gxy # inverse
|
||||
j, k = ((gxy % 1 < g) & (gxy > 1)).T
|
||||
l, m = ((gxi % 1 < g) & (gxi > 1)).T
|
||||
j = torch.stack((torch.ones_like(j), j, k, l, m))
|
||||
t = t.repeat((5, 1, 1))[j]
|
||||
offsets = (torch.zeros_like(gxy)[None] + off[:, None])[j]
|
||||
else:
|
||||
t = targets[0]
|
||||
offsets = 0
|
||||
|
||||
# Define
|
||||
bc, gxy, gwh, a = t.chunk(4, 1) # (image, class), grid xy, grid wh, anchors
|
||||
a, (b, c) = a.long().view(-1), bc.long().T # anchors, image, class
|
||||
gij = (gxy - offsets).long()
|
||||
gi, gj = gij.T # grid indices
|
||||
|
||||
# Append
|
||||
indices.append((b, a, gj.clamp_(0, shape[2] - 1), gi.clamp_(0, shape[3] - 1))) # image, anchor, grid
|
||||
tbox.append(torch.cat((gxy - gij, gwh), 1)) # box
|
||||
anch.append(anchors[a]) # anchors
|
||||
tcls.append(c) # class
|
||||
|
||||
return tcls, tbox, indices, anch
|
||||
@@ -1,381 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Model validation metrics."""
|
||||
|
||||
import math
|
||||
import warnings
|
||||
from pathlib import Path
|
||||
|
||||
import matplotlib.pyplot as plt
|
||||
import numpy as np
|
||||
import torch
|
||||
|
||||
from utils.yolov5.utils import TryExcept, threaded
|
||||
|
||||
|
||||
def fitness(x):
|
||||
"""Calculates fitness of a model using weighted sum of metrics P, R, mAP@0.5, mAP@0.5:0.95."""
|
||||
w = [0.0, 0.0, 0.1, 0.9] # weights for [P, R, mAP@0.5, mAP@0.5:0.95]
|
||||
return (x[:, :4] * w).sum(1)
|
||||
|
||||
|
||||
def smooth(y, f=0.05):
|
||||
"""Applies box filter smoothing to array `y` with fraction `f`, yielding a smoothed array."""
|
||||
nf = round(len(y) * f * 2) // 2 + 1 # number of filter elements (must be odd)
|
||||
p = np.ones(nf // 2) # ones padding
|
||||
yp = np.concatenate((p * y[0], y, p * y[-1]), 0) # y padded
|
||||
return np.convolve(yp, np.ones(nf) / nf, mode="valid") # y-smoothed
|
||||
|
||||
|
||||
def ap_per_class(tp, conf, pred_cls, target_cls, plot=False, save_dir=".", names=(), eps=1e-16, prefix=""):
|
||||
"""
|
||||
Compute the average precision, given the recall and precision curves.
|
||||
|
||||
Source: https://github.com/rafaelpadilla/Object-Detection-Metrics.
|
||||
# Arguments
|
||||
tp: True positives (nparray, nx1 or nx10).
|
||||
conf: Objectness value from 0-1 (nparray).
|
||||
pred_cls: Predicted object classes (nparray).
|
||||
target_cls: True object classes (nparray).
|
||||
plot: Plot precision-recall curve at mAP@0.5
|
||||
save_dir: Plot save directory
|
||||
# Returns
|
||||
The average precision as computed in py-faster-rcnn.
|
||||
"""
|
||||
# Sort by objectness
|
||||
i = np.argsort(-conf)
|
||||
tp, conf, pred_cls = tp[i], conf[i], pred_cls[i]
|
||||
|
||||
# Find unique classes
|
||||
unique_classes, nt = np.unique(target_cls, return_counts=True)
|
||||
nc = unique_classes.shape[0] # number of classes, number of detections
|
||||
|
||||
# Create Precision-Recall curve and compute AP for each class
|
||||
px, py = np.linspace(0, 1, 1000), [] # for plotting
|
||||
ap, p, r = np.zeros((nc, tp.shape[1])), np.zeros((nc, 1000)), np.zeros((nc, 1000))
|
||||
for ci, c in enumerate(unique_classes):
|
||||
i = pred_cls == c
|
||||
n_l = nt[ci] # number of labels
|
||||
n_p = i.sum() # number of predictions
|
||||
if n_p == 0 or n_l == 0:
|
||||
continue
|
||||
|
||||
# Accumulate FPs and TPs
|
||||
fpc = (1 - tp[i]).cumsum(0)
|
||||
tpc = tp[i].cumsum(0)
|
||||
|
||||
# Recall
|
||||
recall = tpc / (n_l + eps) # recall curve
|
||||
r[ci] = np.interp(-px, -conf[i], recall[:, 0], left=0) # negative x, xp because xp decreases
|
||||
|
||||
# Precision
|
||||
precision = tpc / (tpc + fpc) # precision curve
|
||||
p[ci] = np.interp(-px, -conf[i], precision[:, 0], left=1) # p at pr_score
|
||||
|
||||
# AP from recall-precision curve
|
||||
for j in range(tp.shape[1]):
|
||||
ap[ci, j], mpre, mrec = compute_ap(recall[:, j], precision[:, j])
|
||||
if plot and j == 0:
|
||||
py.append(np.interp(px, mrec, mpre)) # precision at mAP@0.5
|
||||
|
||||
# Compute F1 (harmonic mean of precision and recall)
|
||||
f1 = 2 * p * r / (p + r + eps)
|
||||
names = [v for k, v in names.items() if k in unique_classes] # list: only classes that have data
|
||||
names = dict(enumerate(names)) # to dict
|
||||
if plot:
|
||||
plot_pr_curve(px, py, ap, Path(save_dir) / f"{prefix}PR_curve.png", names)
|
||||
plot_mc_curve(px, f1, Path(save_dir) / f"{prefix}F1_curve.png", names, ylabel="F1")
|
||||
plot_mc_curve(px, p, Path(save_dir) / f"{prefix}P_curve.png", names, ylabel="Precision")
|
||||
plot_mc_curve(px, r, Path(save_dir) / f"{prefix}R_curve.png", names, ylabel="Recall")
|
||||
|
||||
i = smooth(f1.mean(0), 0.1).argmax() # max F1 index
|
||||
p, r, f1 = p[:, i], r[:, i], f1[:, i]
|
||||
tp = (r * nt).round() # true positives
|
||||
fp = (tp / (p + eps) - tp).round() # false positives
|
||||
return tp, fp, p, r, f1, ap, unique_classes.astype(int)
|
||||
|
||||
|
||||
def compute_ap(recall, precision):
|
||||
"""Compute the average precision, given the recall and precision curves
|
||||
# Arguments
|
||||
recall: The recall curve (list)
|
||||
precision: The precision curve (list)
|
||||
# Returns
|
||||
Average precision, precision curve, recall curve.
|
||||
"""
|
||||
# Append sentinel values to beginning and end
|
||||
mrec = np.concatenate(([0.0], recall, [1.0]))
|
||||
mpre = np.concatenate(([1.0], precision, [0.0]))
|
||||
|
||||
# Compute the precision envelope
|
||||
mpre = np.flip(np.maximum.accumulate(np.flip(mpre)))
|
||||
|
||||
# Integrate area under curve
|
||||
method = "interp" # methods: 'continuous', 'interp'
|
||||
if method == "interp":
|
||||
x = np.linspace(0, 1, 101) # 101-point interp (COCO)
|
||||
ap = np.trapz(np.interp(x, mrec, mpre), x) # integrate
|
||||
else: # 'continuous'
|
||||
i = np.where(mrec[1:] != mrec[:-1])[0] # points where x axis (recall) changes
|
||||
ap = np.sum((mrec[i + 1] - mrec[i]) * mpre[i + 1]) # area under curve
|
||||
|
||||
return ap, mpre, mrec
|
||||
|
||||
|
||||
class ConfusionMatrix:
|
||||
"""Generates and visualizes a confusion matrix for evaluating object detection classification performance."""
|
||||
|
||||
def __init__(self, nc, conf=0.25, iou_thres=0.45):
|
||||
"""Initializes ConfusionMatrix with given number of classes, confidence, and IoU threshold."""
|
||||
self.matrix = np.zeros((nc + 1, nc + 1))
|
||||
self.nc = nc # number of classes
|
||||
self.conf = conf
|
||||
self.iou_thres = iou_thres
|
||||
|
||||
def process_batch(self, detections, labels):
|
||||
"""
|
||||
Return intersection-over-union (Jaccard index) of boxes.
|
||||
|
||||
Both sets of boxes are expected to be in (x1, y1, x2, y2) format.
|
||||
|
||||
Arguments:
|
||||
detections (Array[N, 6]), x1, y1, x2, y2, conf, class
|
||||
labels (Array[M, 5]), class, x1, y1, x2, y2
|
||||
Returns:
|
||||
None, updates confusion matrix accordingly
|
||||
"""
|
||||
if detections is None:
|
||||
gt_classes = labels.int()
|
||||
for gc in gt_classes:
|
||||
self.matrix[self.nc, gc] += 1 # background FN
|
||||
return
|
||||
|
||||
detections = detections[detections[:, 4] > self.conf]
|
||||
gt_classes = labels[:, 0].int()
|
||||
detection_classes = detections[:, 5].int()
|
||||
iou = box_iou(labels[:, 1:], detections[:, :4])
|
||||
|
||||
x = torch.where(iou > self.iou_thres)
|
||||
if x[0].shape[0]:
|
||||
matches = torch.cat((torch.stack(x, 1), iou[x[0], x[1]][:, None]), 1).cpu().numpy()
|
||||
if x[0].shape[0] > 1:
|
||||
matches = matches[matches[:, 2].argsort()[::-1]]
|
||||
matches = matches[np.unique(matches[:, 1], return_index=True)[1]]
|
||||
matches = matches[matches[:, 2].argsort()[::-1]]
|
||||
matches = matches[np.unique(matches[:, 0], return_index=True)[1]]
|
||||
else:
|
||||
matches = np.zeros((0, 3))
|
||||
|
||||
n = matches.shape[0] > 0
|
||||
m0, m1, _ = matches.transpose().astype(int)
|
||||
for i, gc in enumerate(gt_classes):
|
||||
j = m0 == i
|
||||
if n and sum(j) == 1:
|
||||
self.matrix[detection_classes[m1[j]], gc] += 1 # correct
|
||||
else:
|
||||
self.matrix[self.nc, gc] += 1 # true background
|
||||
|
||||
if n:
|
||||
for i, dc in enumerate(detection_classes):
|
||||
if not any(m1 == i):
|
||||
self.matrix[dc, self.nc] += 1 # predicted background
|
||||
|
||||
def tp_fp(self):
|
||||
"""Calculates true positives (tp) and false positives (fp) excluding the background class from the confusion
|
||||
matrix.
|
||||
"""
|
||||
tp = self.matrix.diagonal() # true positives
|
||||
fp = self.matrix.sum(1) - tp # false positives
|
||||
# fn = self.matrix.sum(0) - tp # false negatives (missed detections)
|
||||
return tp[:-1], fp[:-1] # remove background class
|
||||
|
||||
@TryExcept("WARNING ⚠️ ConfusionMatrix plot failure")
|
||||
def plot(self, normalize=True, save_dir="", names=()):
|
||||
"""Plots confusion matrix using seaborn, optional normalization; can save plot to specified directory."""
|
||||
import seaborn as sn
|
||||
|
||||
array = self.matrix / ((self.matrix.sum(0).reshape(1, -1) + 1e-9) if normalize else 1) # normalize columns
|
||||
array[array < 0.005] = np.nan # don't annotate (would appear as 0.00)
|
||||
|
||||
fig, ax = plt.subplots(1, 1, figsize=(12, 9), tight_layout=True)
|
||||
nc, nn = self.nc, len(names) # number of classes, names
|
||||
sn.set(font_scale=1.0 if nc < 50 else 0.8) # for label size
|
||||
labels = (0 < nn < 99) and (nn == nc) # apply names to ticklabels
|
||||
ticklabels = (names + ["background"]) if labels else "auto"
|
||||
with warnings.catch_warnings():
|
||||
warnings.simplefilter("ignore") # suppress empty matrix RuntimeWarning: All-NaN slice encountered
|
||||
sn.heatmap(
|
||||
array,
|
||||
ax=ax,
|
||||
annot=nc < 30,
|
||||
annot_kws={"size": 8},
|
||||
cmap="Blues",
|
||||
fmt=".2f",
|
||||
square=True,
|
||||
vmin=0.0,
|
||||
xticklabels=ticklabels,
|
||||
yticklabels=ticklabels,
|
||||
).set_facecolor((1, 1, 1))
|
||||
ax.set_xlabel("True")
|
||||
ax.set_ylabel("Predicted")
|
||||
ax.set_title("Confusion Matrix")
|
||||
fig.savefig(Path(save_dir) / "confusion_matrix.png", dpi=250)
|
||||
plt.close(fig)
|
||||
|
||||
def print(self):
|
||||
"""Prints the confusion matrix row-wise, with each class and its predictions separated by spaces."""
|
||||
for i in range(self.nc + 1):
|
||||
print(" ".join(map(str, self.matrix[i])))
|
||||
|
||||
|
||||
def bbox_iou(box1, box2, xywh=True, GIoU=False, DIoU=False, CIoU=False, eps=1e-7):
|
||||
"""
|
||||
Calculates IoU, GIoU, DIoU, or CIoU between two boxes, supporting xywh/xyxy formats.
|
||||
|
||||
Input shapes are box1(1,4) to box2(n,4).
|
||||
"""
|
||||
# Get the coordinates of bounding boxes
|
||||
if xywh: # transform from xywh to xyxy
|
||||
(x1, y1, w1, h1), (x2, y2, w2, h2) = box1.chunk(4, -1), box2.chunk(4, -1)
|
||||
w1_, h1_, w2_, h2_ = w1 / 2, h1 / 2, w2 / 2, h2 / 2
|
||||
b1_x1, b1_x2, b1_y1, b1_y2 = x1 - w1_, x1 + w1_, y1 - h1_, y1 + h1_
|
||||
b2_x1, b2_x2, b2_y1, b2_y2 = x2 - w2_, x2 + w2_, y2 - h2_, y2 + h2_
|
||||
else: # x1, y1, x2, y2 = box1
|
||||
b1_x1, b1_y1, b1_x2, b1_y2 = box1.chunk(4, -1)
|
||||
b2_x1, b2_y1, b2_x2, b2_y2 = box2.chunk(4, -1)
|
||||
w1, h1 = b1_x2 - b1_x1, (b1_y2 - b1_y1).clamp(eps)
|
||||
w2, h2 = b2_x2 - b2_x1, (b2_y2 - b2_y1).clamp(eps)
|
||||
|
||||
# Intersection area
|
||||
inter = (b1_x2.minimum(b2_x2) - b1_x1.maximum(b2_x1)).clamp(0) * (
|
||||
b1_y2.minimum(b2_y2) - b1_y1.maximum(b2_y1)
|
||||
).clamp(0)
|
||||
|
||||
# Union Area
|
||||
union = w1 * h1 + w2 * h2 - inter + eps
|
||||
|
||||
# IoU
|
||||
iou = inter / union
|
||||
if CIoU or DIoU or GIoU:
|
||||
cw = b1_x2.maximum(b2_x2) - b1_x1.minimum(b2_x1) # convex (smallest enclosing box) width
|
||||
ch = b1_y2.maximum(b2_y2) - b1_y1.minimum(b2_y1) # convex height
|
||||
if CIoU or DIoU: # Distance or Complete IoU https://arxiv.org/abs/1911.08287v1
|
||||
c2 = cw**2 + ch**2 + eps # convex diagonal squared
|
||||
rho2 = ((b2_x1 + b2_x2 - b1_x1 - b1_x2) ** 2 + (b2_y1 + b2_y2 - b1_y1 - b1_y2) ** 2) / 4 # center dist ** 2
|
||||
if CIoU: # https://github.com/Zzh-tju/DIoU-SSD-pytorch/blob/master/utils/box/box_utils.py#L47
|
||||
v = (4 / math.pi**2) * (torch.atan(w2 / h2) - torch.atan(w1 / h1)).pow(2)
|
||||
with torch.no_grad():
|
||||
alpha = v / (v - iou + (1 + eps))
|
||||
return iou - (rho2 / c2 + v * alpha) # CIoU
|
||||
return iou - rho2 / c2 # DIoU
|
||||
c_area = cw * ch + eps # convex area
|
||||
return iou - (c_area - union) / c_area # GIoU https://arxiv.org/pdf/1902.09630.pdf
|
||||
return iou # IoU
|
||||
|
||||
|
||||
def box_iou(box1, box2, eps=1e-7):
|
||||
# https://github.com/pytorch/vision/blob/master/torchvision/ops/boxes.py
|
||||
"""
|
||||
Return intersection-over-union (Jaccard index) of boxes.
|
||||
|
||||
Both sets of boxes are expected to be in (x1, y1, x2, y2) format.
|
||||
|
||||
Arguments:
|
||||
box1 (Tensor[N, 4])
|
||||
box2 (Tensor[M, 4])
|
||||
|
||||
Returns:
|
||||
iou (Tensor[N, M]): the NxM matrix containing the pairwise
|
||||
IoU values for every element in boxes1 and boxes2
|
||||
"""
|
||||
# inter(N,M) = (rb(N,M,2) - lt(N,M,2)).clamp(0).prod(2)
|
||||
(a1, a2), (b1, b2) = box1.unsqueeze(1).chunk(2, 2), box2.unsqueeze(0).chunk(2, 2)
|
||||
inter = (torch.min(a2, b2) - torch.max(a1, b1)).clamp(0).prod(2)
|
||||
|
||||
# IoU = inter / (area1 + area2 - inter)
|
||||
return inter / ((a2 - a1).prod(2) + (b2 - b1).prod(2) - inter + eps)
|
||||
|
||||
|
||||
def bbox_ioa(box1, box2, eps=1e-7):
|
||||
"""
|
||||
Returns the intersection over box2 area given box1, box2.
|
||||
|
||||
Boxes are x1y1x2y2
|
||||
box1: np.array of shape(4)
|
||||
box2: np.array of shape(nx4)
|
||||
returns: np.array of shape(n)
|
||||
"""
|
||||
# Get the coordinates of bounding boxes
|
||||
b1_x1, b1_y1, b1_x2, b1_y2 = box1
|
||||
b2_x1, b2_y1, b2_x2, b2_y2 = box2.T
|
||||
|
||||
# Intersection area
|
||||
inter_area = (np.minimum(b1_x2, b2_x2) - np.maximum(b1_x1, b2_x1)).clip(0) * (
|
||||
np.minimum(b1_y2, b2_y2) - np.maximum(b1_y1, b2_y1)
|
||||
).clip(0)
|
||||
|
||||
# box2 area
|
||||
box2_area = (b2_x2 - b2_x1) * (b2_y2 - b2_y1) + eps
|
||||
|
||||
# Intersection over box2 area
|
||||
return inter_area / box2_area
|
||||
|
||||
|
||||
def wh_iou(wh1, wh2, eps=1e-7):
|
||||
"""Calculates the Intersection over Union (IoU) for two sets of widths and heights; `wh1` and `wh2` should be nx2
|
||||
and mx2 tensors.
|
||||
"""
|
||||
wh1 = wh1[:, None] # [N,1,2]
|
||||
wh2 = wh2[None] # [1,M,2]
|
||||
inter = torch.min(wh1, wh2).prod(2) # [N,M]
|
||||
return inter / (wh1.prod(2) + wh2.prod(2) - inter + eps) # iou = inter / (area1 + area2 - inter)
|
||||
|
||||
|
||||
# Plots ----------------------------------------------------------------------------------------------------------------
|
||||
|
||||
|
||||
@threaded
|
||||
def plot_pr_curve(px, py, ap, save_dir=Path("pr_curve.png"), names=()):
|
||||
"""Plots precision-recall curve, optionally per class, saving to `save_dir`; `px`, `py` are lists, `ap` is Nx2
|
||||
array, `names` optional.
|
||||
"""
|
||||
fig, ax = plt.subplots(1, 1, figsize=(9, 6), tight_layout=True)
|
||||
py = np.stack(py, axis=1)
|
||||
|
||||
if 0 < len(names) < 21: # display per-class legend if < 21 classes
|
||||
for i, y in enumerate(py.T):
|
||||
ax.plot(px, y, linewidth=1, label=f"{names[i]} {ap[i, 0]:.3f}") # plot(recall, precision)
|
||||
else:
|
||||
ax.plot(px, py, linewidth=1, color="grey") # plot(recall, precision)
|
||||
|
||||
ax.plot(px, py.mean(1), linewidth=3, color="blue", label=f"all classes {ap[:, 0].mean():.3f} mAP@0.5")
|
||||
ax.set_xlabel("Recall")
|
||||
ax.set_ylabel("Precision")
|
||||
ax.set_xlim(0, 1)
|
||||
ax.set_ylim(0, 1)
|
||||
ax.legend(bbox_to_anchor=(1.04, 1), loc="upper left")
|
||||
ax.set_title("Precision-Recall Curve")
|
||||
fig.savefig(save_dir, dpi=250)
|
||||
plt.close(fig)
|
||||
|
||||
|
||||
@threaded
|
||||
def plot_mc_curve(px, py, save_dir=Path("mc_curve.png"), names=(), xlabel="Confidence", ylabel="Metric"):
|
||||
"""Plots a metric-confidence curve for model predictions, supporting per-class visualization and smoothing."""
|
||||
fig, ax = plt.subplots(1, 1, figsize=(9, 6), tight_layout=True)
|
||||
|
||||
if 0 < len(names) < 21: # display per-class legend if < 21 classes
|
||||
for i, y in enumerate(py):
|
||||
ax.plot(px, y, linewidth=1, label=f"{names[i]}") # plot(confidence, metric)
|
||||
else:
|
||||
ax.plot(px, py.T, linewidth=1, color="grey") # plot(confidence, metric)
|
||||
|
||||
y = smooth(py.mean(0), 0.05)
|
||||
ax.plot(px, y, linewidth=3, color="blue", label=f"all classes {y.max():.2f} at {px[y.argmax()]:.3f}")
|
||||
ax.set_xlabel(xlabel)
|
||||
ax.set_ylabel(ylabel)
|
||||
ax.set_xlim(0, 1)
|
||||
ax.set_ylim(0, 1)
|
||||
ax.legend(bbox_to_anchor=(1.04, 1), loc="upper left")
|
||||
ax.set_title(f"{ylabel}-Confidence Curve")
|
||||
fig.savefig(save_dir, dpi=250)
|
||||
plt.close(fig)
|
||||
@@ -1,517 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Plotting utils."""
|
||||
|
||||
import contextlib
|
||||
import math
|
||||
import os
|
||||
from copy import copy
|
||||
from pathlib import Path
|
||||
|
||||
import cv2
|
||||
import matplotlib
|
||||
import matplotlib.pyplot as plt
|
||||
import numpy as np
|
||||
import pandas as pd
|
||||
import seaborn as sn
|
||||
import torch
|
||||
from PIL import Image, ImageDraw
|
||||
from scipy.ndimage.filters import gaussian_filter1d
|
||||
from ultralytics.utils.plotting import Annotator
|
||||
|
||||
from utils.yolov5.utils import TryExcept, threaded
|
||||
from utils.yolov5.utils.general import LOGGER, clip_boxes, increment_path, xywh2xyxy, xyxy2xywh
|
||||
from utils.yolov5.utils.metrics import fitness
|
||||
|
||||
# Settings
|
||||
RANK = int(os.getenv("RANK", -1))
|
||||
matplotlib.rc("font", **{"size": 11})
|
||||
matplotlib.use("Agg") # for writing to files only
|
||||
|
||||
|
||||
class Colors:
|
||||
"""Provides an RGB color palette derived from Ultralytics color scheme for visualization tasks."""
|
||||
|
||||
def __init__(self):
|
||||
"""
|
||||
Initializes the Colors class with a palette derived from Ultralytics color scheme, converting hex codes to RGB.
|
||||
|
||||
Colors derived from `hex = matplotlib.colors.TABLEAU_COLORS.values()`.
|
||||
"""
|
||||
hexs = (
|
||||
"FF3838",
|
||||
"FF9D97",
|
||||
"FF701F",
|
||||
"FFB21D",
|
||||
"CFD231",
|
||||
"48F90A",
|
||||
"92CC17",
|
||||
"3DDB86",
|
||||
"1A9334",
|
||||
"00D4BB",
|
||||
"2C99A8",
|
||||
"00C2FF",
|
||||
"344593",
|
||||
"6473FF",
|
||||
"0018EC",
|
||||
"8438FF",
|
||||
"520085",
|
||||
"CB38FF",
|
||||
"FF95C8",
|
||||
"FF37C7",
|
||||
)
|
||||
self.palette = [self.hex2rgb(f"#{c}") for c in hexs]
|
||||
self.n = len(self.palette)
|
||||
|
||||
def __call__(self, i, bgr=False):
|
||||
"""Returns color from palette by index `i`, in BGR format if `bgr=True`, else RGB; `i` is an integer index."""
|
||||
c = self.palette[int(i) % self.n]
|
||||
return (c[2], c[1], c[0]) if bgr else c
|
||||
|
||||
@staticmethod
|
||||
def hex2rgb(h):
|
||||
"""Converts hexadecimal color `h` to an RGB tuple (PIL-compatible) with order (R, G, B)."""
|
||||
return tuple(int(h[1 + i : 1 + i + 2], 16) for i in (0, 2, 4))
|
||||
|
||||
|
||||
colors = Colors() # create instance for 'from utils.plots import colors'
|
||||
|
||||
|
||||
def feature_visualization(x, module_type, stage, n=32, save_dir=Path("runs/detect/exp")):
|
||||
"""
|
||||
x: Features to be visualized
|
||||
module_type: Module type
|
||||
stage: Module stage within model
|
||||
n: Maximum number of feature maps to plot
|
||||
save_dir: Directory to save results.
|
||||
"""
|
||||
if ("Detect" not in module_type) and (
|
||||
"Segment" not in module_type
|
||||
): # 'Detect' for Object Detect task,'Segment' for Segment task
|
||||
batch, channels, height, width = x.shape # batch, channels, height, width
|
||||
if height > 1 and width > 1:
|
||||
f = save_dir / f"stage{stage}_{module_type.split('.')[-1]}_features.png" # filename
|
||||
|
||||
blocks = torch.chunk(x[0].cpu(), channels, dim=0) # select batch index 0, block by channels
|
||||
n = min(n, channels) # number of plots
|
||||
fig, ax = plt.subplots(math.ceil(n / 8), 8, tight_layout=True) # 8 rows x n/8 cols
|
||||
ax = ax.ravel()
|
||||
plt.subplots_adjust(wspace=0.05, hspace=0.05)
|
||||
for i in range(n):
|
||||
ax[i].imshow(blocks[i].squeeze()) # cmap='gray'
|
||||
ax[i].axis("off")
|
||||
|
||||
LOGGER.info(f"Saving {f}... ({n}/{channels})")
|
||||
plt.savefig(f, dpi=300, bbox_inches="tight")
|
||||
plt.close()
|
||||
np.save(str(f.with_suffix(".npy")), x[0].cpu().numpy()) # npy save
|
||||
|
||||
|
||||
def hist2d(x, y, n=100):
|
||||
"""
|
||||
Generates a logarithmic 2D histogram, useful for visualizing label or evolution distributions.
|
||||
|
||||
Used in used in labels.png and evolve.png.
|
||||
"""
|
||||
xedges, yedges = np.linspace(x.min(), x.max(), n), np.linspace(y.min(), y.max(), n)
|
||||
hist, xedges, yedges = np.histogram2d(x, y, (xedges, yedges))
|
||||
xidx = np.clip(np.digitize(x, xedges) - 1, 0, hist.shape[0] - 1)
|
||||
yidx = np.clip(np.digitize(y, yedges) - 1, 0, hist.shape[1] - 1)
|
||||
return np.log(hist[xidx, yidx])
|
||||
|
||||
|
||||
def butter_lowpass_filtfilt(data, cutoff=1500, fs=50000, order=5):
|
||||
"""Applies a low-pass Butterworth filter to `data` with specified `cutoff`, `fs`, and `order`."""
|
||||
from scipy.signal import butter, filtfilt
|
||||
|
||||
# https://stackoverflow.com/questions/28536191/how-to-filter-smooth-with-scipy-numpy
|
||||
def butter_lowpass(cutoff, fs, order):
|
||||
"""Applies a low-pass Butterworth filter to a signal with specified cutoff frequency, sample rate, and filter
|
||||
order.
|
||||
"""
|
||||
nyq = 0.5 * fs
|
||||
normal_cutoff = cutoff / nyq
|
||||
return butter(order, normal_cutoff, btype="low", analog=False)
|
||||
|
||||
b, a = butter_lowpass(cutoff, fs, order=order)
|
||||
return filtfilt(b, a, data) # forward-backward filter
|
||||
|
||||
|
||||
def output_to_target(output, max_det=300):
|
||||
"""Converts YOLOv5 model output to [batch_id, class_id, x, y, w, h, conf] format for plotting, limiting detections
|
||||
to `max_det`.
|
||||
"""
|
||||
targets = []
|
||||
for i, o in enumerate(output):
|
||||
box, conf, cls = o[:max_det, :6].cpu().split((4, 1, 1), 1)
|
||||
j = torch.full((conf.shape[0], 1), i)
|
||||
targets.append(torch.cat((j, cls, xyxy2xywh(box), conf), 1))
|
||||
return torch.cat(targets, 0).numpy()
|
||||
|
||||
|
||||
@threaded
|
||||
def plot_images(images, targets, paths=None, fname="images.jpg", names=None):
|
||||
"""Plots an image grid with labels from YOLOv5 predictions or targets, saving to `fname`."""
|
||||
if isinstance(images, torch.Tensor):
|
||||
images = images.cpu().float().numpy()
|
||||
if isinstance(targets, torch.Tensor):
|
||||
targets = targets.cpu().numpy()
|
||||
|
||||
max_size = 1920 # max image size
|
||||
max_subplots = 16 # max image subplots, i.e. 4x4
|
||||
bs, _, h, w = images.shape # batch size, _, height, width
|
||||
bs = min(bs, max_subplots) # limit plot images
|
||||
ns = np.ceil(bs**0.5) # number of subplots (square)
|
||||
if np.max(images[0]) <= 1:
|
||||
images *= 255 # de-normalise (optional)
|
||||
|
||||
# Build Image
|
||||
mosaic = np.full((int(ns * h), int(ns * w), 3), 255, dtype=np.uint8) # init
|
||||
for i, im in enumerate(images):
|
||||
if i == max_subplots: # if last batch has fewer images than we expect
|
||||
break
|
||||
x, y = int(w * (i // ns)), int(h * (i % ns)) # block origin
|
||||
im = im.transpose(1, 2, 0)
|
||||
mosaic[y : y + h, x : x + w, :] = im
|
||||
|
||||
# Resize (optional)
|
||||
scale = max_size / ns / max(h, w)
|
||||
if scale < 1:
|
||||
h = math.ceil(scale * h)
|
||||
w = math.ceil(scale * w)
|
||||
mosaic = cv2.resize(mosaic, tuple(int(x * ns) for x in (w, h)))
|
||||
|
||||
# Annotate
|
||||
fs = int((h + w) * ns * 0.01) # font size
|
||||
annotator = Annotator(mosaic, line_width=round(fs / 10), font_size=fs, pil=True, example=names)
|
||||
for i in range(bs):
|
||||
x, y = int(w * (i // ns)), int(h * (i % ns)) # block origin
|
||||
annotator.rectangle([x, y, x + w, y + h], None, (255, 255, 255), width=2) # borders
|
||||
if paths:
|
||||
annotator.text([x + 5, y + 5], text=Path(paths[i]).name[:40], txt_color=(220, 220, 220)) # filenames
|
||||
if len(targets) > 0:
|
||||
ti = targets[targets[:, 0] == i] # image targets
|
||||
boxes = xywh2xyxy(ti[:, 2:6]).T
|
||||
classes = ti[:, 1].astype("int")
|
||||
labels = ti.shape[1] == 6 # labels if no conf column
|
||||
conf = None if labels else ti[:, 6] # check for confidence presence (label vs pred)
|
||||
|
||||
if boxes.shape[1]:
|
||||
if boxes.max() <= 1.01: # if normalized with tolerance 0.01
|
||||
boxes[[0, 2]] *= w # scale to pixels
|
||||
boxes[[1, 3]] *= h
|
||||
elif scale < 1: # absolute coords need scale if image scales
|
||||
boxes *= scale
|
||||
boxes[[0, 2]] += x
|
||||
boxes[[1, 3]] += y
|
||||
for j, box in enumerate(boxes.T.tolist()):
|
||||
cls = classes[j]
|
||||
color = colors(cls)
|
||||
cls = names[cls] if names else cls
|
||||
if labels or conf[j] > 0.25: # 0.25 conf thresh
|
||||
label = f"{cls}" if labels else f"{cls} {conf[j]:.1f}"
|
||||
annotator.box_label(box, label, color=color)
|
||||
annotator.im.save(fname) # save
|
||||
|
||||
|
||||
def plot_lr_scheduler(optimizer, scheduler, epochs=300, save_dir=""):
|
||||
"""Plots learning rate schedule for given optimizer and scheduler, saving plot to `save_dir`."""
|
||||
optimizer, scheduler = copy(optimizer), copy(scheduler) # do not modify originals
|
||||
y = []
|
||||
for _ in range(epochs):
|
||||
scheduler.step()
|
||||
y.append(optimizer.param_groups[0]["lr"])
|
||||
plt.plot(y, ".-", label="LR")
|
||||
plt.xlabel("epoch")
|
||||
plt.ylabel("LR")
|
||||
plt.grid()
|
||||
plt.xlim(0, epochs)
|
||||
plt.ylim(0)
|
||||
plt.savefig(Path(save_dir) / "LR.png", dpi=200)
|
||||
plt.close()
|
||||
|
||||
|
||||
def plot_val_txt():
|
||||
"""
|
||||
Plots 2D and 1D histograms of bounding box centers from 'val.txt' using matplotlib, saving as 'hist2d.png' and
|
||||
'hist1d.png'.
|
||||
|
||||
Example: from utils.plots import *; plot_val()
|
||||
"""
|
||||
x = np.loadtxt("val.txt", dtype=np.float32)
|
||||
box = xyxy2xywh(x[:, :4])
|
||||
cx, cy = box[:, 0], box[:, 1]
|
||||
|
||||
fig, ax = plt.subplots(1, 1, figsize=(6, 6), tight_layout=True)
|
||||
ax.hist2d(cx, cy, bins=600, cmax=10, cmin=0)
|
||||
ax.set_aspect("equal")
|
||||
plt.savefig("hist2d.png", dpi=300)
|
||||
|
||||
fig, ax = plt.subplots(1, 2, figsize=(12, 6), tight_layout=True)
|
||||
ax[0].hist(cx, bins=600)
|
||||
ax[1].hist(cy, bins=600)
|
||||
plt.savefig("hist1d.png", dpi=200)
|
||||
|
||||
|
||||
def plot_targets_txt():
|
||||
"""
|
||||
Plots histograms of object detection targets from 'targets.txt', saving the figure as 'targets.jpg'.
|
||||
|
||||
Example: from utils.plots import *; plot_targets_txt()
|
||||
"""
|
||||
x = np.loadtxt("targets.txt", dtype=np.float32).T
|
||||
s = ["x targets", "y targets", "width targets", "height targets"]
|
||||
fig, ax = plt.subplots(2, 2, figsize=(8, 8), tight_layout=True)
|
||||
ax = ax.ravel()
|
||||
for i in range(4):
|
||||
ax[i].hist(x[i], bins=100, label=f"{x[i].mean():.3g} +/- {x[i].std():.3g}")
|
||||
ax[i].legend()
|
||||
ax[i].set_title(s[i])
|
||||
plt.savefig("targets.jpg", dpi=200)
|
||||
|
||||
|
||||
def plot_val_study(file="", dir="", x=None):
|
||||
"""
|
||||
Plots validation study results from 'study*.txt' files in a directory or a specific file, comparing model
|
||||
performance and speed.
|
||||
|
||||
Example: from utils.plots import *; plot_val_study()
|
||||
"""
|
||||
save_dir = Path(file).parent if file else Path(dir)
|
||||
plot2 = False # plot additional results
|
||||
if plot2:
|
||||
ax = plt.subplots(2, 4, figsize=(10, 6), tight_layout=True)[1].ravel()
|
||||
|
||||
fig2, ax2 = plt.subplots(1, 1, figsize=(8, 4), tight_layout=True)
|
||||
# for f in [save_dir / f'study_coco_{x}.txt' for x in ['yolov5n6', 'yolov5s6', 'yolov5m6', 'yolov5l6', 'yolov5x6']]:
|
||||
for f in sorted(save_dir.glob("study*.txt")):
|
||||
y = np.loadtxt(f, dtype=np.float32, usecols=[0, 1, 2, 3, 7, 8, 9], ndmin=2).T
|
||||
x = np.arange(y.shape[1]) if x is None else np.array(x)
|
||||
if plot2:
|
||||
s = ["P", "R", "mAP@.5", "mAP@.5:.95", "t_preprocess (ms/img)", "t_inference (ms/img)", "t_NMS (ms/img)"]
|
||||
for i in range(7):
|
||||
ax[i].plot(x, y[i], ".-", linewidth=2, markersize=8)
|
||||
ax[i].set_title(s[i])
|
||||
|
||||
j = y[3].argmax() + 1
|
||||
ax2.plot(
|
||||
y[5, 1:j],
|
||||
y[3, 1:j] * 1e2,
|
||||
".-",
|
||||
linewidth=2,
|
||||
markersize=8,
|
||||
label=f.stem.replace("study_coco_", "").replace("yolo", "YOLO"),
|
||||
)
|
||||
|
||||
ax2.plot(
|
||||
1e3 / np.array([209, 140, 97, 58, 35, 18]),
|
||||
[34.6, 40.5, 43.0, 47.5, 49.7, 51.5],
|
||||
"k.-",
|
||||
linewidth=2,
|
||||
markersize=8,
|
||||
alpha=0.25,
|
||||
label="EfficientDet",
|
||||
)
|
||||
|
||||
ax2.grid(alpha=0.2)
|
||||
ax2.set_yticks(np.arange(20, 60, 5))
|
||||
ax2.set_xlim(0, 57)
|
||||
ax2.set_ylim(25, 55)
|
||||
ax2.set_xlabel("GPU Speed (ms/img)")
|
||||
ax2.set_ylabel("COCO AP val")
|
||||
ax2.legend(loc="lower right")
|
||||
f = save_dir / "study.png"
|
||||
print(f"Saving {f}...")
|
||||
plt.savefig(f, dpi=300)
|
||||
|
||||
|
||||
@TryExcept() # known issue https://github.com/ultralytics/yolov5/issues/5395
|
||||
def plot_labels(labels, names=(), save_dir=Path("")):
|
||||
"""Plots dataset labels, saving correlogram and label images, handles classes, and visualizes bounding boxes."""
|
||||
LOGGER.info(f"Plotting labels to {save_dir / 'labels.jpg'}... ")
|
||||
c, b = labels[:, 0], labels[:, 1:].transpose() # classes, boxes
|
||||
nc = int(c.max() + 1) # number of classes
|
||||
x = pd.DataFrame(b.transpose(), columns=["x", "y", "width", "height"])
|
||||
|
||||
# seaborn correlogram
|
||||
sn.pairplot(x, corner=True, diag_kind="auto", kind="hist", diag_kws=dict(bins=50), plot_kws=dict(pmax=0.9))
|
||||
plt.savefig(save_dir / "labels_correlogram.jpg", dpi=200)
|
||||
plt.close()
|
||||
|
||||
# matplotlib labels
|
||||
matplotlib.use("svg") # faster
|
||||
ax = plt.subplots(2, 2, figsize=(8, 8), tight_layout=True)[1].ravel()
|
||||
y = ax[0].hist(c, bins=np.linspace(0, nc, nc + 1) - 0.5, rwidth=0.8)
|
||||
with contextlib.suppress(Exception): # color histogram bars by class
|
||||
[y[2].patches[i].set_color([x / 255 for x in colors(i)]) for i in range(nc)] # known issue #3195
|
||||
ax[0].set_ylabel("instances")
|
||||
if 0 < len(names) < 30:
|
||||
ax[0].set_xticks(range(len(names)))
|
||||
ax[0].set_xticklabels(list(names.values()), rotation=90, fontsize=10)
|
||||
else:
|
||||
ax[0].set_xlabel("classes")
|
||||
sn.histplot(x, x="x", y="y", ax=ax[2], bins=50, pmax=0.9)
|
||||
sn.histplot(x, x="width", y="height", ax=ax[3], bins=50, pmax=0.9)
|
||||
|
||||
# rectangles
|
||||
labels[:, 1:3] = 0.5 # center
|
||||
labels[:, 1:] = xywh2xyxy(labels[:, 1:]) * 2000
|
||||
img = Image.fromarray(np.ones((2000, 2000, 3), dtype=np.uint8) * 255)
|
||||
for cls, *box in labels[:1000]:
|
||||
ImageDraw.Draw(img).rectangle(box, width=1, outline=colors(cls)) # plot
|
||||
ax[1].imshow(img)
|
||||
ax[1].axis("off")
|
||||
|
||||
for a in [0, 1, 2, 3]:
|
||||
for s in ["top", "right", "left", "bottom"]:
|
||||
ax[a].spines[s].set_visible(False)
|
||||
|
||||
plt.savefig(save_dir / "labels.jpg", dpi=200)
|
||||
matplotlib.use("Agg")
|
||||
plt.close()
|
||||
|
||||
|
||||
def imshow_cls(im, labels=None, pred=None, names=None, nmax=25, verbose=False, f=Path("images.jpg")):
|
||||
"""Displays a grid of images with optional labels and predictions, saving to a file."""
|
||||
from utils.yolov5.utils.augmentations import denormalize
|
||||
|
||||
names = names or [f"class{i}" for i in range(1000)]
|
||||
blocks = torch.chunk(
|
||||
denormalize(im.clone()).cpu().float(), len(im), dim=0
|
||||
) # select batch index 0, block by channels
|
||||
n = min(len(blocks), nmax) # number of plots
|
||||
m = min(8, round(n**0.5)) # 8 x 8 default
|
||||
fig, ax = plt.subplots(math.ceil(n / m), m) # 8 rows x n/8 cols
|
||||
ax = ax.ravel() if m > 1 else [ax]
|
||||
# plt.subplots_adjust(wspace=0.05, hspace=0.05)
|
||||
for i in range(n):
|
||||
ax[i].imshow(blocks[i].squeeze().permute((1, 2, 0)).numpy().clip(0.0, 1.0))
|
||||
ax[i].axis("off")
|
||||
if labels is not None:
|
||||
s = names[labels[i]] + (f"—{names[pred[i]]}" if pred is not None else "")
|
||||
ax[i].set_title(s, fontsize=8, verticalalignment="top")
|
||||
plt.savefig(f, dpi=300, bbox_inches="tight")
|
||||
plt.close()
|
||||
if verbose:
|
||||
LOGGER.info(f"Saving {f}")
|
||||
if labels is not None:
|
||||
LOGGER.info("True: " + " ".join(f"{names[i]:3s}" for i in labels[:nmax]))
|
||||
if pred is not None:
|
||||
LOGGER.info("Predicted:" + " ".join(f"{names[i]:3s}" for i in pred[:nmax]))
|
||||
return f
|
||||
|
||||
|
||||
def plot_evolve(evolve_csv="path/to/evolve.csv"):
|
||||
"""
|
||||
Plots hyperparameter evolution results from a given CSV, saving the plot and displaying best results.
|
||||
|
||||
Example: from utils.plots import *; plot_evolve()
|
||||
"""
|
||||
evolve_csv = Path(evolve_csv)
|
||||
data = pd.read_csv(evolve_csv)
|
||||
keys = [x.strip() for x in data.columns]
|
||||
x = data.values
|
||||
f = fitness(x)
|
||||
j = np.argmax(f) # max fitness index
|
||||
plt.figure(figsize=(10, 12), tight_layout=True)
|
||||
matplotlib.rc("font", **{"size": 8})
|
||||
print(f"Best results from row {j} of {evolve_csv}:")
|
||||
for i, k in enumerate(keys[7:]):
|
||||
v = x[:, 7 + i]
|
||||
mu = v[j] # best single result
|
||||
plt.subplot(6, 5, i + 1)
|
||||
plt.scatter(v, f, c=hist2d(v, f, 20), cmap="viridis", alpha=0.8, edgecolors="none")
|
||||
plt.plot(mu, f.max(), "k+", markersize=15)
|
||||
plt.title(f"{k} = {mu:.3g}", fontdict={"size": 9}) # limit to 40 characters
|
||||
if i % 5 != 0:
|
||||
plt.yticks([])
|
||||
print(f"{k:>15}: {mu:.3g}")
|
||||
f = evolve_csv.with_suffix(".png") # filename
|
||||
plt.savefig(f, dpi=200)
|
||||
plt.close()
|
||||
print(f"Saved {f}")
|
||||
|
||||
|
||||
def plot_results(file="path/to/results.csv", dir=""):
|
||||
"""
|
||||
Plots training results from a 'results.csv' file; accepts file path and directory as arguments.
|
||||
|
||||
Example: from utils.plots import *; plot_results('path/to/results.csv')
|
||||
"""
|
||||
save_dir = Path(file).parent if file else Path(dir)
|
||||
fig, ax = plt.subplots(2, 5, figsize=(12, 6), tight_layout=True)
|
||||
ax = ax.ravel()
|
||||
files = list(save_dir.glob("results*.csv"))
|
||||
assert len(files), f"No results.csv files found in {save_dir.resolve()}, nothing to plot."
|
||||
for f in files:
|
||||
try:
|
||||
data = pd.read_csv(f)
|
||||
s = [x.strip() for x in data.columns]
|
||||
x = data.values[:, 0]
|
||||
for i, j in enumerate([1, 2, 3, 4, 5, 8, 9, 10, 6, 7]):
|
||||
y = data.values[:, j].astype("float")
|
||||
# y[y == 0] = np.nan # don't show zero values
|
||||
ax[i].plot(x, y, marker=".", label=f.stem, linewidth=2, markersize=8) # actual results
|
||||
ax[i].plot(x, gaussian_filter1d(y, sigma=3), ":", label="smooth", linewidth=2) # smoothing line
|
||||
ax[i].set_title(s[j], fontsize=12)
|
||||
# if j in [8, 9, 10]: # share train and val loss y axes
|
||||
# ax[i].get_shared_y_axes().join(ax[i], ax[i - 5])
|
||||
except Exception as e:
|
||||
LOGGER.info(f"Warning: Plotting error for {f}: {e}")
|
||||
ax[1].legend()
|
||||
fig.savefig(save_dir / "results.png", dpi=200)
|
||||
plt.close()
|
||||
|
||||
|
||||
def profile_idetection(start=0, stop=0, labels=(), save_dir=""):
|
||||
"""
|
||||
Plots per-image iDetection logs, comparing metrics like storage and performance over time.
|
||||
|
||||
Example: from utils.plots import *; profile_idetection()
|
||||
"""
|
||||
ax = plt.subplots(2, 4, figsize=(12, 6), tight_layout=True)[1].ravel()
|
||||
s = ["Images", "Free Storage (GB)", "RAM Usage (GB)", "Battery", "dt_raw (ms)", "dt_smooth (ms)", "real-world FPS"]
|
||||
files = list(Path(save_dir).glob("frames*.txt"))
|
||||
for fi, f in enumerate(files):
|
||||
try:
|
||||
results = np.loadtxt(f, ndmin=2).T[:, 90:-30] # clip first and last rows
|
||||
n = results.shape[1] # number of rows
|
||||
x = np.arange(start, min(stop, n) if stop else n)
|
||||
results = results[:, x]
|
||||
t = results[0] - results[0].min() # set t0=0s
|
||||
results[0] = x
|
||||
for i, a in enumerate(ax):
|
||||
if i < len(results):
|
||||
label = labels[fi] if len(labels) else f.stem.replace("frames_", "")
|
||||
a.plot(t, results[i], marker=".", label=label, linewidth=1, markersize=5)
|
||||
a.set_title(s[i])
|
||||
a.set_xlabel("time (s)")
|
||||
# if fi == len(files) - 1:
|
||||
# a.set_ylim(bottom=0)
|
||||
for side in ["top", "right"]:
|
||||
a.spines[side].set_visible(False)
|
||||
else:
|
||||
a.remove()
|
||||
except Exception as e:
|
||||
print(f"Warning: Plotting error for {f}; {e}")
|
||||
ax[1].legend()
|
||||
plt.savefig(Path(save_dir) / "idetection_profile.png", dpi=200)
|
||||
|
||||
|
||||
def save_one_box(xyxy, im, file=Path("im.jpg"), gain=1.02, pad=10, square=False, BGR=False, save=True):
|
||||
"""Crops and saves an image from bounding box `xyxy`, applied with `gain` and `pad`, optionally squares and adjusts
|
||||
for BGR.
|
||||
"""
|
||||
xyxy = torch.tensor(xyxy).view(-1, 4)
|
||||
b = xyxy2xywh(xyxy) # boxes
|
||||
if square:
|
||||
b[:, 2:] = b[:, 2:].max(1)[0].unsqueeze(1) # attempt rectangle to square
|
||||
b[:, 2:] = b[:, 2:] * gain + pad # box wh * gain + pad
|
||||
xyxy = xywh2xyxy(b).long()
|
||||
clip_boxes(xyxy, im.shape)
|
||||
crop = im[int(xyxy[0, 1]) : int(xyxy[0, 3]), int(xyxy[0, 0]) : int(xyxy[0, 2]), :: (1 if BGR else -1)]
|
||||
if save:
|
||||
file.parent.mkdir(parents=True, exist_ok=True) # make directory
|
||||
f = str(increment_path(file).with_suffix(".jpg"))
|
||||
# cv2.imwrite(f, crop) # save BGR, https://github.com/ultralytics/yolov5/issues/7007 chroma subsampling issue
|
||||
Image.fromarray(crop[..., ::-1]).save(f, quality=95, subsampling=0) # save RGB
|
||||
return crop
|
||||
@@ -1 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
@@ -1,92 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Image augmentation functions."""
|
||||
|
||||
import math
|
||||
import random
|
||||
|
||||
import cv2
|
||||
import numpy as np
|
||||
|
||||
from ..augmentations import box_candidates
|
||||
from ..general import resample_segments, segment2box
|
||||
|
||||
|
||||
def mixup(im, labels, segments, im2, labels2, segments2):
|
||||
"""
|
||||
Applies MixUp augmentation blending two images, labels, and segments with a random ratio.
|
||||
|
||||
See https://arxiv.org/pdf/1710.09412.pdf
|
||||
"""
|
||||
r = np.random.beta(32.0, 32.0) # mixup ratio, alpha=beta=32.0
|
||||
im = (im * r + im2 * (1 - r)).astype(np.uint8)
|
||||
labels = np.concatenate((labels, labels2), 0)
|
||||
segments = np.concatenate((segments, segments2), 0)
|
||||
return im, labels, segments
|
||||
|
||||
|
||||
def random_perspective(
|
||||
im, targets=(), segments=(), degrees=10, translate=0.1, scale=0.1, shear=10, perspective=0.0, border=(0, 0)
|
||||
):
|
||||
# torchvision.transforms.RandomAffine(degrees=(-10, 10), translate=(.1, .1), scale=(.9, 1.1), shear=(-10, 10))
|
||||
# targets = [cls, xyxy]
|
||||
"""Applies random perspective, rotation, scale, shear, and translation augmentations to an image and targets."""
|
||||
height = im.shape[0] + border[0] * 2 # shape(h,w,c)
|
||||
width = im.shape[1] + border[1] * 2
|
||||
|
||||
# Center
|
||||
C = np.eye(3)
|
||||
C[0, 2] = -im.shape[1] / 2 # x translation (pixels)
|
||||
C[1, 2] = -im.shape[0] / 2 # y translation (pixels)
|
||||
|
||||
# Perspective
|
||||
P = np.eye(3)
|
||||
P[2, 0] = random.uniform(-perspective, perspective) # x perspective (about y)
|
||||
P[2, 1] = random.uniform(-perspective, perspective) # y perspective (about x)
|
||||
|
||||
# Rotation and Scale
|
||||
R = np.eye(3)
|
||||
a = random.uniform(-degrees, degrees)
|
||||
# a += random.choice([-180, -90, 0, 90]) # add 90deg rotations to small rotations
|
||||
s = random.uniform(1 - scale, 1 + scale)
|
||||
# s = 2 ** random.uniform(-scale, scale)
|
||||
R[:2] = cv2.getRotationMatrix2D(angle=a, center=(0, 0), scale=s)
|
||||
|
||||
# Shear
|
||||
S = np.eye(3)
|
||||
S[0, 1] = math.tan(random.uniform(-shear, shear) * math.pi / 180) # x shear (deg)
|
||||
S[1, 0] = math.tan(random.uniform(-shear, shear) * math.pi / 180) # y shear (deg)
|
||||
|
||||
# Translation
|
||||
T = np.eye(3)
|
||||
T[0, 2] = random.uniform(0.5 - translate, 0.5 + translate) * width # x translation (pixels)
|
||||
T[1, 2] = random.uniform(0.5 - translate, 0.5 + translate) * height # y translation (pixels)
|
||||
|
||||
# Combined rotation matrix
|
||||
M = T @ S @ R @ P @ C # order of operations (right to left) is IMPORTANT
|
||||
if (border[0] != 0) or (border[1] != 0) or (M != np.eye(3)).any(): # image changed
|
||||
if perspective:
|
||||
im = cv2.warpPerspective(im, M, dsize=(width, height), borderValue=(114, 114, 114))
|
||||
else: # affine
|
||||
im = cv2.warpAffine(im, M[:2], dsize=(width, height), borderValue=(114, 114, 114))
|
||||
|
||||
new_segments = []
|
||||
if n := len(targets):
|
||||
new = np.zeros((n, 4))
|
||||
segments = resample_segments(segments) # upsample
|
||||
for i, segment in enumerate(segments):
|
||||
xy = np.ones((len(segment), 3))
|
||||
xy[:, :2] = segment
|
||||
xy = xy @ M.T # transform
|
||||
xy = xy[:, :2] / xy[:, 2:3] if perspective else xy[:, :2] # perspective rescale or affine
|
||||
|
||||
# clip
|
||||
new[i] = segment2box(xy, width, height)
|
||||
new_segments.append(xy)
|
||||
|
||||
# filter candidates
|
||||
i = box_candidates(box1=targets[:, 1:5].T * s, box2=new.T, area_thr=0.01)
|
||||
targets = targets[i]
|
||||
targets[:, 1:5] = new[i]
|
||||
new_segments = np.array(new_segments)[i]
|
||||
|
||||
return im, targets, new_segments
|
||||
@@ -1,366 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Dataloaders."""
|
||||
|
||||
import os
|
||||
import random
|
||||
|
||||
import cv2
|
||||
import numpy as np
|
||||
import torch
|
||||
from torch.utils.data import DataLoader
|
||||
|
||||
from ..augmentations import augment_hsv, copy_paste, letterbox
|
||||
from ..dataloaders import InfiniteDataLoader, LoadImagesAndLabels, SmartDistributedSampler, seed_worker
|
||||
from ..general import LOGGER, xyn2xy, xywhn2xyxy, xyxy2xywhn
|
||||
from ..torch_utils import torch_distributed_zero_first
|
||||
from .augmentations import mixup, random_perspective
|
||||
|
||||
RANK = int(os.getenv("RANK", -1))
|
||||
|
||||
|
||||
def create_dataloader(
|
||||
path,
|
||||
imgsz,
|
||||
batch_size,
|
||||
stride,
|
||||
single_cls=False,
|
||||
hyp=None,
|
||||
augment=False,
|
||||
cache=False,
|
||||
pad=0.0,
|
||||
rect=False,
|
||||
rank=-1,
|
||||
workers=8,
|
||||
image_weights=False,
|
||||
quad=False,
|
||||
prefix="",
|
||||
shuffle=False,
|
||||
mask_downsample_ratio=1,
|
||||
overlap_mask=False,
|
||||
seed=0,
|
||||
):
|
||||
"""Creates a dataloader for training, validating, or testing YOLO models with various dataset options."""
|
||||
if rect and shuffle:
|
||||
LOGGER.warning("WARNING ⚠️ --rect is incompatible with DataLoader shuffle, setting shuffle=False")
|
||||
shuffle = False
|
||||
with torch_distributed_zero_first(rank): # init dataset *.cache only once if DDP
|
||||
dataset = LoadImagesAndLabelsAndMasks(
|
||||
path,
|
||||
imgsz,
|
||||
batch_size,
|
||||
augment=augment, # augmentation
|
||||
hyp=hyp, # hyperparameters
|
||||
rect=rect, # rectangular batches
|
||||
cache_images=cache,
|
||||
single_cls=single_cls,
|
||||
stride=int(stride),
|
||||
pad=pad,
|
||||
image_weights=image_weights,
|
||||
prefix=prefix,
|
||||
downsample_ratio=mask_downsample_ratio,
|
||||
overlap=overlap_mask,
|
||||
rank=rank,
|
||||
)
|
||||
|
||||
batch_size = min(batch_size, len(dataset))
|
||||
nd = torch.cuda.device_count() # number of CUDA devices
|
||||
nw = min([os.cpu_count() // max(nd, 1), batch_size if batch_size > 1 else 0, workers]) # number of workers
|
||||
sampler = None if rank == -1 else SmartDistributedSampler(dataset, shuffle=shuffle)
|
||||
loader = DataLoader if image_weights else InfiniteDataLoader # only DataLoader allows for attribute updates
|
||||
generator = torch.Generator()
|
||||
generator.manual_seed(6148914691236517205 + seed + RANK)
|
||||
return loader(
|
||||
dataset,
|
||||
batch_size=batch_size,
|
||||
shuffle=shuffle and sampler is None,
|
||||
num_workers=nw,
|
||||
sampler=sampler,
|
||||
drop_last=quad,
|
||||
pin_memory=True,
|
||||
collate_fn=LoadImagesAndLabelsAndMasks.collate_fn4 if quad else LoadImagesAndLabelsAndMasks.collate_fn,
|
||||
worker_init_fn=seed_worker,
|
||||
generator=generator,
|
||||
), dataset
|
||||
|
||||
|
||||
class LoadImagesAndLabelsAndMasks(LoadImagesAndLabels): # for training/testing
|
||||
"""Loads images, labels, and segmentation masks for training and testing YOLO models with augmentation support."""
|
||||
|
||||
def __init__(
|
||||
self,
|
||||
path,
|
||||
img_size=640,
|
||||
batch_size=16,
|
||||
augment=False,
|
||||
hyp=None,
|
||||
rect=False,
|
||||
image_weights=False,
|
||||
cache_images=False,
|
||||
single_cls=False,
|
||||
stride=32,
|
||||
pad=0,
|
||||
min_items=0,
|
||||
prefix="",
|
||||
downsample_ratio=1,
|
||||
overlap=False,
|
||||
rank=-1,
|
||||
seed=0,
|
||||
):
|
||||
"""Initializes the dataset with image, label, and mask loading capabilities for training/testing."""
|
||||
super().__init__(
|
||||
path,
|
||||
img_size,
|
||||
batch_size,
|
||||
augment,
|
||||
hyp,
|
||||
rect,
|
||||
image_weights,
|
||||
cache_images,
|
||||
single_cls,
|
||||
stride,
|
||||
pad,
|
||||
min_items,
|
||||
prefix,
|
||||
rank,
|
||||
seed,
|
||||
)
|
||||
self.downsample_ratio = downsample_ratio
|
||||
self.overlap = overlap
|
||||
|
||||
def __getitem__(self, index):
|
||||
"""Returns a transformed item from the dataset at the specified index, handling indexing and image weighting."""
|
||||
index = self.indices[index] # linear, shuffled, or image_weights
|
||||
|
||||
hyp = self.hyp
|
||||
if mosaic := self.mosaic and random.random() < hyp["mosaic"]:
|
||||
# Load mosaic
|
||||
img, labels, segments = self.load_mosaic(index)
|
||||
shapes = None
|
||||
|
||||
# MixUp augmentation
|
||||
if random.random() < hyp["mixup"]:
|
||||
img, labels, segments = mixup(img, labels, segments, *self.load_mosaic(random.randint(0, self.n - 1)))
|
||||
|
||||
else:
|
||||
# Load image
|
||||
img, (h0, w0), (h, w) = self.load_image(index)
|
||||
|
||||
# Letterbox
|
||||
shape = self.batch_shapes[self.batch[index]] if self.rect else self.img_size # final letterboxed shape
|
||||
img, ratio, pad = letterbox(img, shape, auto=False, scaleup=self.augment)
|
||||
shapes = (h0, w0), ((h / h0, w / w0), pad) # for COCO mAP rescaling
|
||||
|
||||
labels = self.labels[index].copy()
|
||||
# [array, array, ....], array.shape=(num_points, 2), xyxyxyxy
|
||||
segments = self.segments[index].copy()
|
||||
if len(segments):
|
||||
for i_s in range(len(segments)):
|
||||
segments[i_s] = xyn2xy(
|
||||
segments[i_s],
|
||||
ratio[0] * w,
|
||||
ratio[1] * h,
|
||||
padw=pad[0],
|
||||
padh=pad[1],
|
||||
)
|
||||
if labels.size: # normalized xywh to pixel xyxy format
|
||||
labels[:, 1:] = xywhn2xyxy(labels[:, 1:], ratio[0] * w, ratio[1] * h, padw=pad[0], padh=pad[1])
|
||||
|
||||
if self.augment:
|
||||
img, labels, segments = random_perspective(
|
||||
img,
|
||||
labels,
|
||||
segments=segments,
|
||||
degrees=hyp["degrees"],
|
||||
translate=hyp["translate"],
|
||||
scale=hyp["scale"],
|
||||
shear=hyp["shear"],
|
||||
perspective=hyp["perspective"],
|
||||
)
|
||||
|
||||
nl = len(labels) # number of labels
|
||||
masks = []
|
||||
if nl:
|
||||
labels[:, 1:5] = xyxy2xywhn(labels[:, 1:5], w=img.shape[1], h=img.shape[0], clip=True, eps=1e-3)
|
||||
if self.overlap:
|
||||
masks, sorted_idx = polygons2masks_overlap(
|
||||
img.shape[:2], segments, downsample_ratio=self.downsample_ratio
|
||||
)
|
||||
masks = masks[None] # (640, 640) -> (1, 640, 640)
|
||||
labels = labels[sorted_idx]
|
||||
else:
|
||||
masks = polygons2masks(img.shape[:2], segments, color=1, downsample_ratio=self.downsample_ratio)
|
||||
|
||||
masks = (
|
||||
torch.from_numpy(masks)
|
||||
if len(masks)
|
||||
else torch.zeros(
|
||||
1 if self.overlap else nl, img.shape[0] // self.downsample_ratio, img.shape[1] // self.downsample_ratio
|
||||
)
|
||||
)
|
||||
# TODO: albumentations support
|
||||
if self.augment:
|
||||
# Albumentations
|
||||
# there are some augmentation that won't change boxes and masks,
|
||||
# so just be it for now.
|
||||
img, labels = self.albumentations(img, labels)
|
||||
nl = len(labels) # update after albumentations
|
||||
|
||||
# HSV color-space
|
||||
augment_hsv(img, hgain=hyp["hsv_h"], sgain=hyp["hsv_s"], vgain=hyp["hsv_v"])
|
||||
|
||||
# Flip up-down
|
||||
if random.random() < hyp["flipud"]:
|
||||
img = np.flipud(img)
|
||||
if nl:
|
||||
labels[:, 2] = 1 - labels[:, 2]
|
||||
masks = torch.flip(masks, dims=[1])
|
||||
|
||||
# Flip left-right
|
||||
if random.random() < hyp["fliplr"]:
|
||||
img = np.fliplr(img)
|
||||
if nl:
|
||||
labels[:, 1] = 1 - labels[:, 1]
|
||||
masks = torch.flip(masks, dims=[2])
|
||||
|
||||
# Cutouts # labels = cutout(img, labels, p=0.5)
|
||||
|
||||
labels_out = torch.zeros((nl, 6))
|
||||
if nl:
|
||||
labels_out[:, 1:] = torch.from_numpy(labels)
|
||||
|
||||
# Convert
|
||||
img = img.transpose((2, 0, 1))[::-1] # HWC to CHW, BGR to RGB
|
||||
img = np.ascontiguousarray(img)
|
||||
|
||||
return (torch.from_numpy(img), labels_out, self.im_files[index], shapes, masks)
|
||||
|
||||
def load_mosaic(self, index):
|
||||
"""Loads 1 image + 3 random images into a 4-image YOLOv5 mosaic, adjusting labels and segments accordingly."""
|
||||
labels4, segments4 = [], []
|
||||
s = self.img_size
|
||||
yc, xc = (int(random.uniform(-x, 2 * s + x)) for x in self.mosaic_border) # mosaic center x, y
|
||||
|
||||
# 3 additional image indices
|
||||
indices = [index] + random.choices(self.indices, k=3) # 3 additional image indices
|
||||
for i, index in enumerate(indices):
|
||||
# Load image
|
||||
img, _, (h, w) = self.load_image(index)
|
||||
|
||||
# place img in img4
|
||||
if i == 0: # top left
|
||||
img4 = np.full((s * 2, s * 2, img.shape[2]), 114, dtype=np.uint8) # base image with 4 tiles
|
||||
x1a, y1a, x2a, y2a = max(xc - w, 0), max(yc - h, 0), xc, yc # xmin, ymin, xmax, ymax (large image)
|
||||
x1b, y1b, x2b, y2b = w - (x2a - x1a), h - (y2a - y1a), w, h # xmin, ymin, xmax, ymax (small image)
|
||||
elif i == 1: # top right
|
||||
x1a, y1a, x2a, y2a = xc, max(yc - h, 0), min(xc + w, s * 2), yc
|
||||
x1b, y1b, x2b, y2b = 0, h - (y2a - y1a), min(w, x2a - x1a), h
|
||||
elif i == 2: # bottom left
|
||||
x1a, y1a, x2a, y2a = max(xc - w, 0), yc, xc, min(s * 2, yc + h)
|
||||
x1b, y1b, x2b, y2b = w - (x2a - x1a), 0, w, min(y2a - y1a, h)
|
||||
elif i == 3: # bottom right
|
||||
x1a, y1a, x2a, y2a = xc, yc, min(xc + w, s * 2), min(s * 2, yc + h)
|
||||
x1b, y1b, x2b, y2b = 0, 0, min(w, x2a - x1a), min(y2a - y1a, h)
|
||||
|
||||
img4[y1a:y2a, x1a:x2a] = img[y1b:y2b, x1b:x2b] # img4[ymin:ymax, xmin:xmax]
|
||||
padw = x1a - x1b
|
||||
padh = y1a - y1b
|
||||
|
||||
labels, segments = self.labels[index].copy(), self.segments[index].copy()
|
||||
|
||||
if labels.size:
|
||||
labels[:, 1:] = xywhn2xyxy(labels[:, 1:], w, h, padw, padh) # normalized xywh to pixel xyxy format
|
||||
segments = [xyn2xy(x, w, h, padw, padh) for x in segments]
|
||||
labels4.append(labels)
|
||||
segments4.extend(segments)
|
||||
|
||||
# Concat/clip labels
|
||||
labels4 = np.concatenate(labels4, 0)
|
||||
for x in (labels4[:, 1:], *segments4):
|
||||
np.clip(x, 0, 2 * s, out=x) # clip when using random_perspective()
|
||||
# img4, labels4 = replicate(img4, labels4) # replicate
|
||||
|
||||
# Augment
|
||||
img4, labels4, segments4 = copy_paste(img4, labels4, segments4, p=self.hyp["copy_paste"])
|
||||
img4, labels4, segments4 = random_perspective(
|
||||
img4,
|
||||
labels4,
|
||||
segments4,
|
||||
degrees=self.hyp["degrees"],
|
||||
translate=self.hyp["translate"],
|
||||
scale=self.hyp["scale"],
|
||||
shear=self.hyp["shear"],
|
||||
perspective=self.hyp["perspective"],
|
||||
border=self.mosaic_border,
|
||||
) # border to remove
|
||||
return img4, labels4, segments4
|
||||
|
||||
@staticmethod
|
||||
def collate_fn(batch):
|
||||
"""Custom collation function for DataLoader, batches images, labels, paths, shapes, and segmentation masks."""
|
||||
img, label, path, shapes, masks = zip(*batch) # transposed
|
||||
batched_masks = torch.cat(masks, 0)
|
||||
for i, l in enumerate(label):
|
||||
l[:, 0] = i # add target image index for build_targets()
|
||||
return torch.stack(img, 0), torch.cat(label, 0), path, shapes, batched_masks
|
||||
|
||||
|
||||
def polygon2mask(img_size, polygons, color=1, downsample_ratio=1):
|
||||
"""
|
||||
Args:
|
||||
img_size (tuple): The image size.
|
||||
polygons (np.ndarray): [N, M], N is the number of polygons,
|
||||
M is the number of points(Be divided by 2).
|
||||
"""
|
||||
mask = np.zeros(img_size, dtype=np.uint8)
|
||||
polygons = np.asarray(polygons)
|
||||
polygons = polygons.astype(np.int32)
|
||||
shape = polygons.shape
|
||||
polygons = polygons.reshape(shape[0], -1, 2)
|
||||
cv2.fillPoly(mask, polygons, color=color)
|
||||
nh, nw = (img_size[0] // downsample_ratio, img_size[1] // downsample_ratio)
|
||||
# NOTE: fillPoly firstly then resize is trying the keep the same way
|
||||
# of loss calculation when mask-ratio=1.
|
||||
mask = cv2.resize(mask, (nw, nh))
|
||||
return mask
|
||||
|
||||
|
||||
def polygons2masks(img_size, polygons, color, downsample_ratio=1):
|
||||
"""
|
||||
Args:
|
||||
img_size (tuple): The image size.
|
||||
polygons (list[np.ndarray]): each polygon is [N, M],
|
||||
N is the number of polygons,
|
||||
M is the number of points(Be divided by 2).
|
||||
"""
|
||||
masks = []
|
||||
for si in range(len(polygons)):
|
||||
mask = polygon2mask(img_size, [polygons[si].reshape(-1)], color, downsample_ratio)
|
||||
masks.append(mask)
|
||||
return np.array(masks)
|
||||
|
||||
|
||||
def polygons2masks_overlap(img_size, segments, downsample_ratio=1):
|
||||
"""Return a (640, 640) overlap mask."""
|
||||
masks = np.zeros(
|
||||
(img_size[0] // downsample_ratio, img_size[1] // downsample_ratio),
|
||||
dtype=np.int32 if len(segments) > 255 else np.uint8,
|
||||
)
|
||||
areas = []
|
||||
ms = []
|
||||
for si in range(len(segments)):
|
||||
mask = polygon2mask(
|
||||
img_size,
|
||||
[segments[si].reshape(-1)],
|
||||
downsample_ratio=downsample_ratio,
|
||||
color=1,
|
||||
)
|
||||
ms.append(mask)
|
||||
areas.append(mask.sum())
|
||||
areas = np.asarray(areas)
|
||||
index = np.argsort(-areas)
|
||||
ms = np.array(ms)[index]
|
||||
for i in range(len(segments)):
|
||||
mask = ms[i] * (i + 1)
|
||||
masks = masks + mask
|
||||
masks = np.clip(masks, a_min=0, a_max=i + 1)
|
||||
return masks, index
|
||||
@@ -1,160 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
import cv2
|
||||
import numpy as np
|
||||
import torch
|
||||
import torch.nn.functional as F
|
||||
|
||||
|
||||
def crop_mask(masks, boxes):
|
||||
"""
|
||||
"Crop" predicted masks by zeroing out everything not in the predicted bbox. Vectorized by Chong (thanks Chong).
|
||||
|
||||
Args:
|
||||
- masks should be a size [n, h, w] tensor of masks
|
||||
- boxes should be a size [n, 4] tensor of bbox coords in relative point form
|
||||
"""
|
||||
n, h, w = masks.shape
|
||||
x1, y1, x2, y2 = torch.chunk(boxes[:, :, None], 4, 1) # x1 shape(1,1,n)
|
||||
r = torch.arange(w, device=masks.device, dtype=x1.dtype)[None, None, :] # rows shape(1,w,1)
|
||||
c = torch.arange(h, device=masks.device, dtype=x1.dtype)[None, :, None] # cols shape(h,1,1)
|
||||
|
||||
return masks * ((r >= x1) * (r < x2) * (c >= y1) * (c < y2))
|
||||
|
||||
|
||||
def process_mask_upsample(protos, masks_in, bboxes, shape):
|
||||
"""
|
||||
Crop after upsample.
|
||||
protos: [mask_dim, mask_h, mask_w]
|
||||
masks_in: [n, mask_dim], n is number of masks after nms
|
||||
bboxes: [n, 4], n is number of masks after nms
|
||||
shape: input_image_size, (h, w).
|
||||
|
||||
return: h, w, n
|
||||
"""
|
||||
c, mh, mw = protos.shape # CHW
|
||||
masks = (masks_in @ protos.float().view(c, -1)).sigmoid().view(-1, mh, mw)
|
||||
masks = F.interpolate(masks[None], shape, mode="bilinear", align_corners=False)[0] # CHW
|
||||
masks = crop_mask(masks, bboxes) # CHW
|
||||
return masks.gt_(0.5)
|
||||
|
||||
|
||||
def process_mask(protos, masks_in, bboxes, shape, upsample=False):
|
||||
"""
|
||||
Crop before upsample.
|
||||
proto_out: [mask_dim, mask_h, mask_w]
|
||||
out_masks: [n, mask_dim], n is number of masks after nms
|
||||
bboxes: [n, 4], n is number of masks after nms
|
||||
shape:input_image_size, (h, w).
|
||||
|
||||
return: h, w, n
|
||||
"""
|
||||
c, mh, mw = protos.shape # CHW
|
||||
ih, iw = shape
|
||||
masks = (masks_in @ protos.float().view(c, -1)).sigmoid().view(-1, mh, mw) # CHW
|
||||
|
||||
downsampled_bboxes = bboxes.clone()
|
||||
downsampled_bboxes[:, 0] *= mw / iw
|
||||
downsampled_bboxes[:, 2] *= mw / iw
|
||||
downsampled_bboxes[:, 3] *= mh / ih
|
||||
downsampled_bboxes[:, 1] *= mh / ih
|
||||
|
||||
masks = crop_mask(masks, downsampled_bboxes) # CHW
|
||||
if upsample:
|
||||
masks = F.interpolate(masks[None], shape, mode="bilinear", align_corners=False)[0] # CHW
|
||||
return masks.gt_(0.5)
|
||||
|
||||
|
||||
def process_mask_native(protos, masks_in, bboxes, shape):
|
||||
"""
|
||||
Crop after upsample.
|
||||
protos: [mask_dim, mask_h, mask_w]
|
||||
masks_in: [n, mask_dim], n is number of masks after nms
|
||||
bboxes: [n, 4], n is number of masks after nms
|
||||
shape: input_image_size, (h, w).
|
||||
|
||||
return: h, w, n
|
||||
"""
|
||||
c, mh, mw = protos.shape # CHW
|
||||
masks = (masks_in @ protos.float().view(c, -1)).sigmoid().view(-1, mh, mw)
|
||||
gain = min(mh / shape[0], mw / shape[1]) # gain = old / new
|
||||
pad = (mw - shape[1] * gain) / 2, (mh - shape[0] * gain) / 2 # wh padding
|
||||
top, left = int(pad[1]), int(pad[0]) # y, x
|
||||
bottom, right = int(mh - pad[1]), int(mw - pad[0])
|
||||
masks = masks[:, top:bottom, left:right]
|
||||
|
||||
masks = F.interpolate(masks[None], shape, mode="bilinear", align_corners=False)[0] # CHW
|
||||
masks = crop_mask(masks, bboxes) # CHW
|
||||
return masks.gt_(0.5)
|
||||
|
||||
|
||||
def scale_image(im1_shape, masks, im0_shape, ratio_pad=None):
|
||||
"""
|
||||
img1_shape: model input shape, [h, w]
|
||||
img0_shape: origin pic shape, [h, w, 3]
|
||||
masks: [h, w, num].
|
||||
"""
|
||||
# Rescale coordinates (xyxy) from im1_shape to im0_shape
|
||||
if ratio_pad is None: # calculate from im0_shape
|
||||
gain = min(im1_shape[0] / im0_shape[0], im1_shape[1] / im0_shape[1]) # gain = old / new
|
||||
pad = (im1_shape[1] - im0_shape[1] * gain) / 2, (im1_shape[0] - im0_shape[0] * gain) / 2 # wh padding
|
||||
else:
|
||||
pad = ratio_pad[1]
|
||||
top, left = int(pad[1]), int(pad[0]) # y, x
|
||||
bottom, right = int(im1_shape[0] - pad[1]), int(im1_shape[1] - pad[0])
|
||||
|
||||
if len(masks.shape) < 2:
|
||||
raise ValueError(f'"len of masks shape" should be 2 or 3, but got {len(masks.shape)}')
|
||||
masks = masks[top:bottom, left:right]
|
||||
# masks = masks.permute(2, 0, 1).contiguous()
|
||||
# masks = F.interpolate(masks[None], im0_shape[:2], mode='bilinear', align_corners=False)[0]
|
||||
# masks = masks.permute(1, 2, 0).contiguous()
|
||||
masks = cv2.resize(masks, (im0_shape[1], im0_shape[0]))
|
||||
|
||||
if len(masks.shape) == 2:
|
||||
masks = masks[:, :, None]
|
||||
return masks
|
||||
|
||||
|
||||
def mask_iou(mask1, mask2, eps=1e-7):
|
||||
"""
|
||||
mask1: [N, n] m1 means number of predicted objects
|
||||
mask2: [M, n] m2 means number of gt objects
|
||||
Note: n means image_w x image_h.
|
||||
|
||||
return: masks iou, [N, M]
|
||||
"""
|
||||
intersection = torch.matmul(mask1, mask2.t()).clamp(0)
|
||||
union = (mask1.sum(1)[:, None] + mask2.sum(1)[None]) - intersection # (area1 + area2) - intersection
|
||||
return intersection / (union + eps)
|
||||
|
||||
|
||||
def masks_iou(mask1, mask2, eps=1e-7):
|
||||
"""
|
||||
mask1: [N, n] m1 means number of predicted objects
|
||||
mask2: [N, n] m2 means number of gt objects
|
||||
Note: n means image_w x image_h.
|
||||
|
||||
return: masks iou, (N, )
|
||||
"""
|
||||
intersection = (mask1 * mask2).sum(1).clamp(0) # (N, )
|
||||
union = (mask1.sum(1) + mask2.sum(1))[None] - intersection # (area1 + area2) - intersection
|
||||
return intersection / (union + eps)
|
||||
|
||||
|
||||
def masks2segments(masks, strategy="largest"):
|
||||
"""Converts binary (n,160,160) masks to polygon segments with options for concatenation or selecting the largest
|
||||
segment.
|
||||
"""
|
||||
segments = []
|
||||
for x in masks.int().cpu().numpy().astype("uint8"):
|
||||
c = cv2.findContours(x, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)[0]
|
||||
if c:
|
||||
if strategy == "concat": # concatenate all segments
|
||||
c = np.concatenate([x.reshape(-1, 2) for x in c])
|
||||
elif strategy == "largest": # select largest segment
|
||||
c = np.array(c[np.array([len(x) for x in c]).argmax()]).reshape(-1, 2)
|
||||
else:
|
||||
c = np.zeros((0, 2)) # no segments found
|
||||
segments.append(c.astype("float32"))
|
||||
return segments
|
||||
@@ -1,197 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
import torch
|
||||
import torch.nn as nn
|
||||
import torch.nn.functional as F
|
||||
|
||||
from ..general import xywh2xyxy
|
||||
from ..loss import FocalLoss, smooth_BCE
|
||||
from ..metrics import bbox_iou
|
||||
from ..torch_utils import de_parallel
|
||||
from .general import crop_mask
|
||||
|
||||
|
||||
class ComputeLoss:
|
||||
"""Computes the YOLOv5 model's loss components including classification, objectness, box, and mask losses."""
|
||||
|
||||
def __init__(self, model, autobalance=False, overlap=False):
|
||||
"""Initializes the compute loss function for YOLOv5 models with options for autobalancing and overlap
|
||||
handling.
|
||||
"""
|
||||
self.sort_obj_iou = False
|
||||
self.overlap = overlap
|
||||
device = next(model.parameters()).device # get model device
|
||||
h = model.hyp # hyperparameters
|
||||
|
||||
# Define criteria
|
||||
BCEcls = nn.BCEWithLogitsLoss(pos_weight=torch.tensor([h["cls_pw"]], device=device))
|
||||
BCEobj = nn.BCEWithLogitsLoss(pos_weight=torch.tensor([h["obj_pw"]], device=device))
|
||||
|
||||
# Class label smoothing https://arxiv.org/pdf/1902.04103.pdf eqn 3
|
||||
self.cp, self.cn = smooth_BCE(eps=h.get("label_smoothing", 0.0)) # positive, negative BCE targets
|
||||
|
||||
# Focal loss
|
||||
g = h["fl_gamma"] # focal loss gamma
|
||||
if g > 0:
|
||||
BCEcls, BCEobj = FocalLoss(BCEcls, g), FocalLoss(BCEobj, g)
|
||||
|
||||
m = de_parallel(model).model[-1] # Detect() module
|
||||
self.balance = {3: [4.0, 1.0, 0.4]}.get(m.nl, [4.0, 1.0, 0.25, 0.06, 0.02]) # P3-P7
|
||||
self.ssi = list(m.stride).index(16) if autobalance else 0 # stride 16 index
|
||||
self.BCEcls, self.BCEobj, self.gr, self.hyp, self.autobalance = BCEcls, BCEobj, 1.0, h, autobalance
|
||||
self.na = m.na # number of anchors
|
||||
self.nc = m.nc # number of classes
|
||||
self.nl = m.nl # number of layers
|
||||
self.nm = m.nm # number of masks
|
||||
self.anchors = m.anchors
|
||||
self.device = device
|
||||
|
||||
def __call__(self, preds, targets, masks): # predictions, targets, model
|
||||
"""Evaluates YOLOv5 model's loss for given predictions, targets, and masks; returns total loss components."""
|
||||
p, proto = preds
|
||||
bs, nm, mask_h, mask_w = proto.shape # batch size, number of masks, mask height, mask width
|
||||
lcls = torch.zeros(1, device=self.device)
|
||||
lbox = torch.zeros(1, device=self.device)
|
||||
lobj = torch.zeros(1, device=self.device)
|
||||
lseg = torch.zeros(1, device=self.device)
|
||||
tcls, tbox, indices, anchors, tidxs, xywhn = self.build_targets(p, targets) # targets
|
||||
|
||||
# Losses
|
||||
for i, pi in enumerate(p): # layer index, layer predictions
|
||||
b, a, gj, gi = indices[i] # image, anchor, gridy, gridx
|
||||
tobj = torch.zeros(pi.shape[:4], dtype=pi.dtype, device=self.device) # target obj
|
||||
|
||||
if n := b.shape[0]:
|
||||
pxy, pwh, _, pcls, pmask = pi[b, a, gj, gi].split((2, 2, 1, self.nc, nm), 1) # subset of predictions
|
||||
|
||||
# Box regression
|
||||
pxy = pxy.sigmoid() * 2 - 0.5
|
||||
pwh = (pwh.sigmoid() * 2) ** 2 * anchors[i]
|
||||
pbox = torch.cat((pxy, pwh), 1) # predicted box
|
||||
iou = bbox_iou(pbox, tbox[i], CIoU=True).squeeze() # iou(prediction, target)
|
||||
lbox += (1.0 - iou).mean() # iou loss
|
||||
|
||||
# Objectness
|
||||
iou = iou.detach().clamp(0).type(tobj.dtype)
|
||||
if self.sort_obj_iou:
|
||||
j = iou.argsort()
|
||||
b, a, gj, gi, iou = b[j], a[j], gj[j], gi[j], iou[j]
|
||||
if self.gr < 1:
|
||||
iou = (1.0 - self.gr) + self.gr * iou
|
||||
tobj[b, a, gj, gi] = iou # iou ratio
|
||||
|
||||
# Classification
|
||||
if self.nc > 1: # cls loss (only if multiple classes)
|
||||
t = torch.full_like(pcls, self.cn, device=self.device) # targets
|
||||
t[range(n), tcls[i]] = self.cp
|
||||
lcls += self.BCEcls(pcls, t) # BCE
|
||||
|
||||
# Mask regression
|
||||
if tuple(masks.shape[-2:]) != (mask_h, mask_w): # downsample
|
||||
masks = F.interpolate(masks[None], (mask_h, mask_w), mode="nearest")[0]
|
||||
marea = xywhn[i][:, 2:].prod(1) # mask width, height normalized
|
||||
mxyxy = xywh2xyxy(xywhn[i] * torch.tensor([mask_w, mask_h, mask_w, mask_h], device=self.device))
|
||||
for bi in b.unique():
|
||||
j = b == bi # matching index
|
||||
if self.overlap:
|
||||
mask_gti = torch.where(masks[bi][None] == tidxs[i][j].view(-1, 1, 1), 1.0, 0.0)
|
||||
else:
|
||||
mask_gti = masks[tidxs[i]][j]
|
||||
lseg += self.single_mask_loss(mask_gti, pmask[j], proto[bi], mxyxy[j], marea[j])
|
||||
|
||||
obji = self.BCEobj(pi[..., 4], tobj)
|
||||
lobj += obji * self.balance[i] # obj loss
|
||||
if self.autobalance:
|
||||
self.balance[i] = self.balance[i] * 0.9999 + 0.0001 / obji.detach().item()
|
||||
|
||||
if self.autobalance:
|
||||
self.balance = [x / self.balance[self.ssi] for x in self.balance]
|
||||
lbox *= self.hyp["box"]
|
||||
lobj *= self.hyp["obj"]
|
||||
lcls *= self.hyp["cls"]
|
||||
lseg *= self.hyp["box"] / bs
|
||||
|
||||
loss = lbox + lobj + lcls + lseg
|
||||
return loss * bs, torch.cat((lbox, lseg, lobj, lcls)).detach()
|
||||
|
||||
def single_mask_loss(self, gt_mask, pred, proto, xyxy, area):
|
||||
"""Calculates and normalizes single mask loss for YOLOv5 between predicted and ground truth masks."""
|
||||
pred_mask = (pred @ proto.view(self.nm, -1)).view(-1, *proto.shape[1:]) # (n,32) @ (32,80,80) -> (n,80,80)
|
||||
loss = F.binary_cross_entropy_with_logits(pred_mask, gt_mask, reduction="none")
|
||||
return (crop_mask(loss, xyxy).mean(dim=(1, 2)) / area).mean()
|
||||
|
||||
def build_targets(self, p, targets):
|
||||
"""Prepares YOLOv5 targets for loss computation; inputs targets (image, class, x, y, w, h), output target
|
||||
classes/boxes.
|
||||
"""
|
||||
na, nt = self.na, targets.shape[0] # number of anchors, targets
|
||||
tcls, tbox, indices, anch, tidxs, xywhn = [], [], [], [], [], []
|
||||
gain = torch.ones(8, device=self.device) # normalized to gridspace gain
|
||||
ai = torch.arange(na, device=self.device).float().view(na, 1).repeat(1, nt) # same as .repeat_interleave(nt)
|
||||
if self.overlap:
|
||||
batch = p[0].shape[0]
|
||||
ti = []
|
||||
for i in range(batch):
|
||||
num = (targets[:, 0] == i).sum() # find number of targets of each image
|
||||
ti.append(torch.arange(num, device=self.device).float().view(1, num).repeat(na, 1) + 1) # (na, num)
|
||||
ti = torch.cat(ti, 1) # (na, nt)
|
||||
else:
|
||||
ti = torch.arange(nt, device=self.device).float().view(1, nt).repeat(na, 1)
|
||||
targets = torch.cat((targets.repeat(na, 1, 1), ai[..., None], ti[..., None]), 2) # append anchor indices
|
||||
|
||||
g = 0.5 # bias
|
||||
off = (
|
||||
torch.tensor(
|
||||
[
|
||||
[0, 0],
|
||||
[1, 0],
|
||||
[0, 1],
|
||||
[-1, 0],
|
||||
[0, -1], # j,k,l,m
|
||||
# [1, 1], [1, -1], [-1, 1], [-1, -1], # jk,jm,lk,lm
|
||||
],
|
||||
device=self.device,
|
||||
).float()
|
||||
* g
|
||||
) # offsets
|
||||
|
||||
for i in range(self.nl):
|
||||
anchors, shape = self.anchors[i], p[i].shape
|
||||
gain[2:6] = torch.tensor(shape)[[3, 2, 3, 2]] # xyxy gain
|
||||
|
||||
# Match targets to anchors
|
||||
t = targets * gain # shape(3,n,7)
|
||||
if nt:
|
||||
# Matches
|
||||
r = t[..., 4:6] / anchors[:, None] # wh ratio
|
||||
j = torch.max(r, 1 / r).max(2)[0] < self.hyp["anchor_t"] # compare
|
||||
# j = wh_iou(anchors, t[:, 4:6]) > model.hyp['iou_t'] # iou(3,n)=wh_iou(anchors(3,2), gwh(n,2))
|
||||
t = t[j] # filter
|
||||
|
||||
# Offsets
|
||||
gxy = t[:, 2:4] # grid xy
|
||||
gxi = gain[[2, 3]] - gxy # inverse
|
||||
j, k = ((gxy % 1 < g) & (gxy > 1)).T
|
||||
l, m = ((gxi % 1 < g) & (gxi > 1)).T
|
||||
j = torch.stack((torch.ones_like(j), j, k, l, m))
|
||||
t = t.repeat((5, 1, 1))[j]
|
||||
offsets = (torch.zeros_like(gxy)[None] + off[:, None])[j]
|
||||
else:
|
||||
t = targets[0]
|
||||
offsets = 0
|
||||
|
||||
# Define
|
||||
bc, gxy, gwh, at = t.chunk(4, 1) # (image, class), grid xy, grid wh, anchors
|
||||
(a, tidx), (b, c) = at.long().T, bc.long().T # anchors, image, class
|
||||
gij = (gxy - offsets).long()
|
||||
gi, gj = gij.T # grid indices
|
||||
|
||||
# Append
|
||||
indices.append((b, a, gj.clamp_(0, shape[2] - 1), gi.clamp_(0, shape[3] - 1))) # image, anchor, grid
|
||||
tbox.append(torch.cat((gxy - gij, gwh), 1)) # box
|
||||
anch.append(anchors[a]) # anchors
|
||||
tcls.append(c) # class
|
||||
tidxs.append(tidx)
|
||||
xywhn.append(torch.cat((gxy, gwh), 1) / gain[2:6]) # xywh normalized
|
||||
|
||||
return tcls, tbox, indices, anch, tidxs, xywhn
|
||||
@@ -1,225 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Model validation metrics."""
|
||||
|
||||
import numpy as np
|
||||
|
||||
from ..metrics import ap_per_class
|
||||
|
||||
|
||||
def fitness(x):
|
||||
"""Evaluates model fitness by a weighted sum of 8 metrics, `x`: [N,8] array, weights: [0.1, 0.9] for mAP and F1."""
|
||||
w = [0.0, 0.0, 0.1, 0.9, 0.0, 0.0, 0.1, 0.9]
|
||||
return (x[:, :8] * w).sum(1)
|
||||
|
||||
|
||||
def ap_per_class_box_and_mask(
|
||||
tp_m,
|
||||
tp_b,
|
||||
conf,
|
||||
pred_cls,
|
||||
target_cls,
|
||||
plot=False,
|
||||
save_dir=".",
|
||||
names=(),
|
||||
):
|
||||
"""
|
||||
Args:
|
||||
tp_b: tp of boxes.
|
||||
tp_m: tp of masks.
|
||||
other arguments see `func: ap_per_class`.
|
||||
"""
|
||||
results_boxes = ap_per_class(
|
||||
tp_b, conf, pred_cls, target_cls, plot=plot, save_dir=save_dir, names=names, prefix="Box"
|
||||
)[2:]
|
||||
results_masks = ap_per_class(
|
||||
tp_m, conf, pred_cls, target_cls, plot=plot, save_dir=save_dir, names=names, prefix="Mask"
|
||||
)[2:]
|
||||
|
||||
return {
|
||||
"boxes": {
|
||||
"p": results_boxes[0],
|
||||
"r": results_boxes[1],
|
||||
"ap": results_boxes[3],
|
||||
"f1": results_boxes[2],
|
||||
"ap_class": results_boxes[4],
|
||||
},
|
||||
"masks": {
|
||||
"p": results_masks[0],
|
||||
"r": results_masks[1],
|
||||
"ap": results_masks[3],
|
||||
"f1": results_masks[2],
|
||||
"ap_class": results_masks[4],
|
||||
},
|
||||
}
|
||||
|
||||
|
||||
class Metric:
|
||||
"""Computes performance metrics like precision, recall, F1 score, and average precision for model evaluation."""
|
||||
|
||||
def __init__(self) -> None:
|
||||
"""Initializes performance metric attributes for precision, recall, F1 score, average precision, and class
|
||||
indices.
|
||||
"""
|
||||
self.p = [] # (nc, )
|
||||
self.r = [] # (nc, )
|
||||
self.f1 = [] # (nc, )
|
||||
self.all_ap = [] # (nc, 10)
|
||||
self.ap_class_index = [] # (nc, )
|
||||
|
||||
@property
|
||||
def ap50(self):
|
||||
"""
|
||||
AP@0.5 of all classes.
|
||||
|
||||
Return:
|
||||
(nc, ) or [].
|
||||
"""
|
||||
return self.all_ap[:, 0] if len(self.all_ap) else []
|
||||
|
||||
@property
|
||||
def ap(self):
|
||||
"""AP@0.5:0.95
|
||||
Return:
|
||||
(nc, ) or [].
|
||||
"""
|
||||
return self.all_ap.mean(1) if len(self.all_ap) else []
|
||||
|
||||
@property
|
||||
def mp(self):
|
||||
"""
|
||||
Mean precision of all classes.
|
||||
|
||||
Return:
|
||||
float.
|
||||
"""
|
||||
return self.p.mean() if len(self.p) else 0.0
|
||||
|
||||
@property
|
||||
def mr(self):
|
||||
"""
|
||||
Mean recall of all classes.
|
||||
|
||||
Return:
|
||||
float.
|
||||
"""
|
||||
return self.r.mean() if len(self.r) else 0.0
|
||||
|
||||
@property
|
||||
def map50(self):
|
||||
"""
|
||||
Mean AP@0.5 of all classes.
|
||||
|
||||
Return:
|
||||
float.
|
||||
"""
|
||||
return self.all_ap[:, 0].mean() if len(self.all_ap) else 0.0
|
||||
|
||||
@property
|
||||
def map(self):
|
||||
"""
|
||||
Mean AP@0.5:0.95 of all classes.
|
||||
|
||||
Return:
|
||||
float.
|
||||
"""
|
||||
return self.all_ap.mean() if len(self.all_ap) else 0.0
|
||||
|
||||
def mean_results(self):
|
||||
"""Mean of results, return mp, mr, map50, map."""
|
||||
return (self.mp, self.mr, self.map50, self.map)
|
||||
|
||||
def class_result(self, i):
|
||||
"""Class-aware result, return p[i], r[i], ap50[i], ap[i]."""
|
||||
return (self.p[i], self.r[i], self.ap50[i], self.ap[i])
|
||||
|
||||
def get_maps(self, nc):
|
||||
"""Calculates and returns mean Average Precision (mAP) for each class given number of classes `nc`."""
|
||||
maps = np.zeros(nc) + self.map
|
||||
for i, c in enumerate(self.ap_class_index):
|
||||
maps[c] = self.ap[i]
|
||||
return maps
|
||||
|
||||
def update(self, results):
|
||||
"""
|
||||
Args:
|
||||
results: tuple(p, r, ap, f1, ap_class).
|
||||
"""
|
||||
p, r, all_ap, f1, ap_class_index = results
|
||||
self.p = p
|
||||
self.r = r
|
||||
self.all_ap = all_ap
|
||||
self.f1 = f1
|
||||
self.ap_class_index = ap_class_index
|
||||
|
||||
|
||||
class Metrics:
|
||||
"""Metric for boxes and masks."""
|
||||
|
||||
def __init__(self) -> None:
|
||||
"""Initializes Metric objects for bounding boxes and masks to compute performance metrics in the Metrics
|
||||
class.
|
||||
"""
|
||||
self.metric_box = Metric()
|
||||
self.metric_mask = Metric()
|
||||
|
||||
def update(self, results):
|
||||
"""
|
||||
Args:
|
||||
results: Dict{'boxes': Dict{}, 'masks': Dict{}}.
|
||||
"""
|
||||
self.metric_box.update(list(results["boxes"].values()))
|
||||
self.metric_mask.update(list(results["masks"].values()))
|
||||
|
||||
def mean_results(self):
|
||||
"""Computes and returns the mean results for both box and mask metrics by summing their individual means."""
|
||||
return self.metric_box.mean_results() + self.metric_mask.mean_results()
|
||||
|
||||
def class_result(self, i):
|
||||
"""Returns the sum of box and mask metric results for a specified class index `i`."""
|
||||
return self.metric_box.class_result(i) + self.metric_mask.class_result(i)
|
||||
|
||||
def get_maps(self, nc):
|
||||
"""Calculates and returns the sum of mean average precisions (mAPs) for both box and mask metrics for `nc`
|
||||
classes.
|
||||
"""
|
||||
return self.metric_box.get_maps(nc) + self.metric_mask.get_maps(nc)
|
||||
|
||||
@property
|
||||
def ap_class_index(self):
|
||||
"""Returns the class index for average precision, shared by both box and mask metrics."""
|
||||
return self.metric_box.ap_class_index
|
||||
|
||||
|
||||
KEYS = [
|
||||
"train/box_loss",
|
||||
"train/seg_loss", # train loss
|
||||
"train/obj_loss",
|
||||
"train/cls_loss",
|
||||
"metrics/precision(B)",
|
||||
"metrics/recall(B)",
|
||||
"metrics/mAP_0.5(B)",
|
||||
"metrics/mAP_0.5:0.95(B)", # metrics
|
||||
"metrics/precision(M)",
|
||||
"metrics/recall(M)",
|
||||
"metrics/mAP_0.5(M)",
|
||||
"metrics/mAP_0.5:0.95(M)", # metrics
|
||||
"val/box_loss",
|
||||
"val/seg_loss", # val loss
|
||||
"val/obj_loss",
|
||||
"val/cls_loss",
|
||||
"x/lr0",
|
||||
"x/lr1",
|
||||
"x/lr2",
|
||||
]
|
||||
|
||||
BEST_KEYS = [
|
||||
"best/epoch",
|
||||
"best/precision(B)",
|
||||
"best/recall(B)",
|
||||
"best/mAP_0.5(B)",
|
||||
"best/mAP_0.5:0.95(B)",
|
||||
"best/precision(M)",
|
||||
"best/recall(M)",
|
||||
"best/mAP_0.5(M)",
|
||||
"best/mAP_0.5:0.95(M)",
|
||||
]
|
||||
@@ -1,152 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
import contextlib
|
||||
import math
|
||||
from pathlib import Path
|
||||
|
||||
import cv2
|
||||
import matplotlib.pyplot as plt
|
||||
import numpy as np
|
||||
import pandas as pd
|
||||
import torch
|
||||
|
||||
from .. import threaded
|
||||
from ..general import xywh2xyxy
|
||||
from ..plots import Annotator, colors
|
||||
|
||||
|
||||
@threaded
|
||||
def plot_images_and_masks(images, targets, masks, paths=None, fname="images.jpg", names=None):
|
||||
"""Plots a grid of images, their labels, and masks with optional resizing and annotations, saving to fname."""
|
||||
if isinstance(images, torch.Tensor):
|
||||
images = images.cpu().float().numpy()
|
||||
if isinstance(targets, torch.Tensor):
|
||||
targets = targets.cpu().numpy()
|
||||
if isinstance(masks, torch.Tensor):
|
||||
masks = masks.cpu().numpy().astype(int)
|
||||
|
||||
max_size = 1920 # max image size
|
||||
max_subplots = 16 # max image subplots, i.e. 4x4
|
||||
bs, _, h, w = images.shape # batch size, _, height, width
|
||||
bs = min(bs, max_subplots) # limit plot images
|
||||
ns = np.ceil(bs**0.5) # number of subplots (square)
|
||||
if np.max(images[0]) <= 1:
|
||||
images *= 255 # de-normalise (optional)
|
||||
|
||||
# Build Image
|
||||
mosaic = np.full((int(ns * h), int(ns * w), 3), 255, dtype=np.uint8) # init
|
||||
for i, im in enumerate(images):
|
||||
if i == max_subplots: # if last batch has fewer images than we expect
|
||||
break
|
||||
x, y = int(w * (i // ns)), int(h * (i % ns)) # block origin
|
||||
im = im.transpose(1, 2, 0)
|
||||
mosaic[y : y + h, x : x + w, :] = im
|
||||
|
||||
# Resize (optional)
|
||||
scale = max_size / ns / max(h, w)
|
||||
if scale < 1:
|
||||
h = math.ceil(scale * h)
|
||||
w = math.ceil(scale * w)
|
||||
mosaic = cv2.resize(mosaic, tuple(int(x * ns) for x in (w, h)))
|
||||
|
||||
# Annotate
|
||||
fs = int((h + w) * ns * 0.01) # font size
|
||||
annotator = Annotator(mosaic, line_width=round(fs / 10), font_size=fs, pil=True, example=names)
|
||||
for i in range(i + 1):
|
||||
x, y = int(w * (i // ns)), int(h * (i % ns)) # block origin
|
||||
annotator.rectangle([x, y, x + w, y + h], None, (255, 255, 255), width=2) # borders
|
||||
if paths:
|
||||
annotator.text([x + 5, y + 5], text=Path(paths[i]).name[:40], txt_color=(220, 220, 220)) # filenames
|
||||
if len(targets) > 0:
|
||||
idx = targets[:, 0] == i
|
||||
ti = targets[idx] # image targets
|
||||
|
||||
boxes = xywh2xyxy(ti[:, 2:6]).T
|
||||
classes = ti[:, 1].astype("int")
|
||||
labels = ti.shape[1] == 6 # labels if no conf column
|
||||
conf = None if labels else ti[:, 6] # check for confidence presence (label vs pred)
|
||||
|
||||
if boxes.shape[1]:
|
||||
if boxes.max() <= 1.01: # if normalized with tolerance 0.01
|
||||
boxes[[0, 2]] *= w # scale to pixels
|
||||
boxes[[1, 3]] *= h
|
||||
elif scale < 1: # absolute coords need scale if image scales
|
||||
boxes *= scale
|
||||
boxes[[0, 2]] += x
|
||||
boxes[[1, 3]] += y
|
||||
for j, box in enumerate(boxes.T.tolist()):
|
||||
cls = classes[j]
|
||||
color = colors(cls)
|
||||
cls = names[cls] if names else cls
|
||||
if labels or conf[j] > 0.25: # 0.25 conf thresh
|
||||
label = f"{cls}" if labels else f"{cls} {conf[j]:.1f}"
|
||||
annotator.box_label(box, label, color=color)
|
||||
|
||||
# Plot masks
|
||||
if len(masks):
|
||||
if masks.max() > 1.0: # mean that masks are overlap
|
||||
image_masks = masks[[i]] # (1, 640, 640)
|
||||
nl = len(ti)
|
||||
index = np.arange(nl).reshape(nl, 1, 1) + 1
|
||||
image_masks = np.repeat(image_masks, nl, axis=0)
|
||||
image_masks = np.where(image_masks == index, 1.0, 0.0)
|
||||
else:
|
||||
image_masks = masks[idx]
|
||||
|
||||
im = np.asarray(annotator.im).copy()
|
||||
for j, box in enumerate(boxes.T.tolist()):
|
||||
if labels or conf[j] > 0.25: # 0.25 conf thresh
|
||||
color = colors(classes[j])
|
||||
mh, mw = image_masks[j].shape
|
||||
if mh != h or mw != w:
|
||||
mask = image_masks[j].astype(np.uint8)
|
||||
mask = cv2.resize(mask, (w, h))
|
||||
mask = mask.astype(bool)
|
||||
else:
|
||||
mask = image_masks[j].astype(bool)
|
||||
with contextlib.suppress(Exception):
|
||||
im[y : y + h, x : x + w, :][mask] = (
|
||||
im[y : y + h, x : x + w, :][mask] * 0.4 + np.array(color) * 0.6
|
||||
)
|
||||
annotator.fromarray(im)
|
||||
annotator.im.save(fname) # save
|
||||
|
||||
|
||||
def plot_results_with_masks(file="path/to/results.csv", dir="", best=True):
|
||||
"""
|
||||
Plots training results from CSV files, plotting best or last result highlights based on `best` parameter.
|
||||
|
||||
Example: from utils.plots import *; plot_results('path/to/results.csv')
|
||||
"""
|
||||
save_dir = Path(file).parent if file else Path(dir)
|
||||
fig, ax = plt.subplots(2, 8, figsize=(18, 6), tight_layout=True)
|
||||
ax = ax.ravel()
|
||||
files = list(save_dir.glob("results*.csv"))
|
||||
assert len(files), f"No results.csv files found in {save_dir.resolve()}, nothing to plot."
|
||||
for f in files:
|
||||
try:
|
||||
data = pd.read_csv(f)
|
||||
index = np.argmax(
|
||||
0.9 * data.values[:, 8] + 0.1 * data.values[:, 7] + 0.9 * data.values[:, 12] + 0.1 * data.values[:, 11]
|
||||
)
|
||||
s = [x.strip() for x in data.columns]
|
||||
x = data.values[:, 0]
|
||||
for i, j in enumerate([1, 2, 3, 4, 5, 6, 9, 10, 13, 14, 15, 16, 7, 8, 11, 12]):
|
||||
y = data.values[:, j]
|
||||
# y[y == 0] = np.nan # don't show zero values
|
||||
ax[i].plot(x, y, marker=".", label=f.stem, linewidth=2, markersize=2)
|
||||
if best:
|
||||
# best
|
||||
ax[i].scatter(index, y[index], color="r", label=f"best:{index}", marker="*", linewidth=3)
|
||||
ax[i].set_title(s[j] + f"\n{round(y[index], 5)}")
|
||||
else:
|
||||
# last
|
||||
ax[i].scatter(x[-1], y[-1], color="r", label="last", marker="*", linewidth=3)
|
||||
ax[i].set_title(s[j] + f"\n{round(y[-1], 5)}")
|
||||
# if j in [8, 9, 10]: # share train and val loss y axes
|
||||
# ax[i].get_shared_y_axes().join(ax[i], ax[i - 5])
|
||||
except Exception as e:
|
||||
print(f"Warning: Plotting error for {f}: {e}")
|
||||
ax[1].legend()
|
||||
fig.savefig(save_dir / "results.png", dpi=200)
|
||||
plt.close()
|
||||
@@ -1,482 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""PyTorch utils."""
|
||||
|
||||
import math
|
||||
import os
|
||||
import platform
|
||||
import subprocess
|
||||
import time
|
||||
import warnings
|
||||
from contextlib import contextmanager
|
||||
from copy import deepcopy
|
||||
from pathlib import Path
|
||||
|
||||
import torch
|
||||
import torch.distributed as dist
|
||||
import torch.nn as nn
|
||||
import torch.nn.functional as F
|
||||
from torch.nn.parallel import DistributedDataParallel as DDP
|
||||
|
||||
from utils.yolov5.utils.general import LOGGER, check_version, colorstr, file_date, git_describe
|
||||
|
||||
LOCAL_RANK = int(os.getenv("LOCAL_RANK", -1)) # https://pytorch.org/docs/stable/elastic/run.html
|
||||
RANK = int(os.getenv("RANK", -1))
|
||||
WORLD_SIZE = int(os.getenv("WORLD_SIZE", 1))
|
||||
|
||||
try:
|
||||
import thop # for FLOPs computation
|
||||
except ImportError:
|
||||
thop = None
|
||||
|
||||
# Suppress PyTorch warnings
|
||||
warnings.filterwarnings("ignore", message="User provided device_type of 'cuda', but CUDA is not available. Disabling")
|
||||
warnings.filterwarnings("ignore", category=UserWarning)
|
||||
|
||||
|
||||
def smart_inference_mode(torch_1_9=check_version(torch.__version__, "1.9.0")):
|
||||
"""Applies torch.inference_mode() if torch>=1.9.0, else torch.no_grad() as a decorator for functions."""
|
||||
|
||||
def decorate(fn):
|
||||
"""Applies torch.inference_mode() if torch>=1.9.0, else torch.no_grad() to the decorated function."""
|
||||
return (torch.inference_mode if torch_1_9 else torch.no_grad)()(fn)
|
||||
|
||||
return decorate
|
||||
|
||||
|
||||
def smartCrossEntropyLoss(label_smoothing=0.0):
|
||||
"""Returns a CrossEntropyLoss with optional label smoothing for torch>=1.10.0; warns if smoothing on lower
|
||||
versions.
|
||||
"""
|
||||
if check_version(torch.__version__, "1.10.0"):
|
||||
return nn.CrossEntropyLoss(label_smoothing=label_smoothing)
|
||||
if label_smoothing > 0:
|
||||
LOGGER.warning(f"WARNING ⚠️ label smoothing {label_smoothing} requires torch>=1.10.0")
|
||||
return nn.CrossEntropyLoss()
|
||||
|
||||
|
||||
def smart_DDP(model):
|
||||
"""Initializes DistributedDataParallel (DDP) for model training, respecting torch version constraints."""
|
||||
assert not check_version(torch.__version__, "1.12.0", pinned=True), (
|
||||
"torch==1.12.0 torchvision==0.13.0 DDP training is not supported due to a known issue. "
|
||||
"Please upgrade or downgrade torch to use DDP. See https://github.com/ultralytics/yolov5/issues/8395"
|
||||
)
|
||||
if check_version(torch.__version__, "1.11.0"):
|
||||
return DDP(model, device_ids=[LOCAL_RANK], output_device=LOCAL_RANK, static_graph=True)
|
||||
else:
|
||||
return DDP(model, device_ids=[LOCAL_RANK], output_device=LOCAL_RANK)
|
||||
|
||||
|
||||
def reshape_classifier_output(model, n=1000):
|
||||
"""Reshapes last layer of model to match class count 'n', supporting Classify, Linear, Sequential types."""
|
||||
from utils.yolov5.models.common import Classify
|
||||
|
||||
name, m = list((model.model if hasattr(model, "model") else model).named_children())[-1] # last module
|
||||
if isinstance(m, Classify): # YOLOv5 Classify() head
|
||||
if m.linear.out_features != n:
|
||||
m.linear = nn.Linear(m.linear.in_features, n)
|
||||
elif isinstance(m, nn.Linear): # ResNet, EfficientNet
|
||||
if m.out_features != n:
|
||||
setattr(model, name, nn.Linear(m.in_features, n))
|
||||
elif isinstance(m, nn.Sequential):
|
||||
types = [type(x) for x in m]
|
||||
if nn.Linear in types:
|
||||
i = len(types) - 1 - types[::-1].index(nn.Linear) # last nn.Linear index
|
||||
if m[i].out_features != n:
|
||||
m[i] = nn.Linear(m[i].in_features, n)
|
||||
elif nn.Conv2d in types:
|
||||
i = len(types) - 1 - types[::-1].index(nn.Conv2d) # last nn.Conv2d index
|
||||
if m[i].out_channels != n:
|
||||
m[i] = nn.Conv2d(m[i].in_channels, n, m[i].kernel_size, m[i].stride, bias=m[i].bias is not None)
|
||||
|
||||
|
||||
@contextmanager
|
||||
def torch_distributed_zero_first(local_rank: int):
|
||||
"""Context manager ensuring ordered operations in distributed training by making all processes wait for the leading
|
||||
process.
|
||||
"""
|
||||
if local_rank not in [-1, 0]:
|
||||
dist.barrier(device_ids=[local_rank])
|
||||
yield
|
||||
if local_rank == 0:
|
||||
dist.barrier(device_ids=[0])
|
||||
|
||||
|
||||
def device_count():
|
||||
"""Returns the number of available CUDA devices; works on Linux and Windows by invoking `nvidia-smi`."""
|
||||
assert platform.system() in ("Linux", "Windows"), "device_count() only supported on Linux or Windows"
|
||||
try:
|
||||
cmd = "nvidia-smi -L | wc -l" if platform.system() == "Linux" else 'nvidia-smi -L | find /c /v ""' # Windows
|
||||
return int(subprocess.run(cmd, shell=True, capture_output=True, check=True).stdout.decode().split()[-1])
|
||||
except Exception:
|
||||
return 0
|
||||
|
||||
|
||||
def select_device(device="", batch_size=0, newline=True):
|
||||
"""Selects computing device (CPU, CUDA GPU, MPS) for YOLOv5 model deployment, logging device info."""
|
||||
s = f"YOLOv5 🚀 {git_describe() or file_date()} Python-{platform.python_version()} torch-{torch.__version__} "
|
||||
device = str(device).strip().lower().replace("cuda:", "").replace("none", "") # to string, 'cuda:0' to '0'
|
||||
cpu = device == "cpu"
|
||||
mps = device == "mps" # Apple Metal Performance Shaders (MPS)
|
||||
if cpu or mps:
|
||||
os.environ["CUDA_VISIBLE_DEVICES"] = "-1" # force torch.cuda.is_available() = False
|
||||
elif device: # non-cpu device requested
|
||||
os.environ["CUDA_VISIBLE_DEVICES"] = device # set environment variable - must be before assert is_available()
|
||||
assert torch.cuda.is_available() and torch.cuda.device_count() >= len(device.replace(",", "")), (
|
||||
f"Invalid CUDA '--device {device}' requested, use '--device cpu' or pass valid CUDA device(s)"
|
||||
)
|
||||
|
||||
if not cpu and not mps and torch.cuda.is_available(): # prefer GPU if available
|
||||
devices = device.split(",") if device else "0" # range(torch.cuda.device_count()) # i.e. 0,1,6,7
|
||||
n = len(devices) # device count
|
||||
if n > 1 and batch_size > 0: # check batch_size is divisible by device_count
|
||||
assert batch_size % n == 0, f"batch-size {batch_size} not multiple of GPU count {n}"
|
||||
space = " " * (len(s) + 1)
|
||||
for i, d in enumerate(devices):
|
||||
p = torch.cuda.get_device_properties(i)
|
||||
s += f"{'' if i == 0 else space}CUDA:{d} ({p.name}, {p.total_memory / (1 << 20):.0f}MiB)\n" # bytes to MB
|
||||
arg = "cuda:0"
|
||||
elif mps and getattr(torch, "has_mps", False) and torch.backends.mps.is_available(): # prefer MPS if available
|
||||
s += "MPS\n"
|
||||
arg = "mps"
|
||||
else: # revert to CPU
|
||||
s += "CPU\n"
|
||||
arg = "cpu"
|
||||
|
||||
if not newline:
|
||||
s = s.rstrip()
|
||||
LOGGER.info(s)
|
||||
return torch.device(arg)
|
||||
|
||||
|
||||
def time_sync():
|
||||
"""Synchronizes PyTorch for accurate timing, leveraging CUDA if available, and returns the current time."""
|
||||
if torch.cuda.is_available():
|
||||
torch.cuda.synchronize()
|
||||
return time.time()
|
||||
|
||||
|
||||
def profile(input, ops, n=10, device=None):
|
||||
"""YOLOv5 speed/memory/FLOPs profiler
|
||||
Usage:
|
||||
input = torch.randn(16, 3, 640, 640)
|
||||
m1 = lambda x: x * torch.sigmoid(x)
|
||||
m2 = nn.SiLU()
|
||||
profile(input, [m1, m2], n=100) # profile over 100 iterations.
|
||||
"""
|
||||
results = []
|
||||
if not isinstance(device, torch.device):
|
||||
device = select_device(device)
|
||||
print(
|
||||
f"{'Params':>12s}{'GFLOPs':>12s}{'GPU_mem (GB)':>14s}{'forward (ms)':>14s}{'backward (ms)':>14s}"
|
||||
f"{'input':>24s}{'output':>24s}"
|
||||
)
|
||||
|
||||
for x in input if isinstance(input, list) else [input]:
|
||||
x = x.to(device)
|
||||
x.requires_grad = True
|
||||
for m in ops if isinstance(ops, list) else [ops]:
|
||||
m = m.to(device) if hasattr(m, "to") else m # device
|
||||
m = m.half() if hasattr(m, "half") and isinstance(x, torch.Tensor) and x.dtype is torch.float16 else m
|
||||
tf, tb, t = 0, 0, [0, 0, 0] # dt forward, backward
|
||||
try:
|
||||
flops = thop.profile(m, inputs=(x,), verbose=False)[0] / 1e9 * 2 # GFLOPs
|
||||
except Exception:
|
||||
flops = 0
|
||||
|
||||
try:
|
||||
for _ in range(n):
|
||||
t[0] = time_sync()
|
||||
y = m(x)
|
||||
t[1] = time_sync()
|
||||
try:
|
||||
_ = (sum(yi.sum() for yi in y) if isinstance(y, list) else y).sum().backward()
|
||||
t[2] = time_sync()
|
||||
except Exception: # no backward method
|
||||
# print(e) # for debug
|
||||
t[2] = float("nan")
|
||||
tf += (t[1] - t[0]) * 1000 / n # ms per op forward
|
||||
tb += (t[2] - t[1]) * 1000 / n # ms per op backward
|
||||
mem = torch.cuda.memory_reserved() / 1e9 if torch.cuda.is_available() else 0 # (GB)
|
||||
s_in, s_out = (tuple(x.shape) if isinstance(x, torch.Tensor) else "list" for x in (x, y)) # shapes
|
||||
p = sum(x.numel() for x in m.parameters()) if isinstance(m, nn.Module) else 0 # parameters
|
||||
print(f"{p:12}{flops:12.4g}{mem:>14.3f}{tf:14.4g}{tb:14.4g}{str(s_in):>24s}{str(s_out):>24s}")
|
||||
results.append([p, flops, mem, tf, tb, s_in, s_out])
|
||||
except Exception as e:
|
||||
print(e)
|
||||
results.append(None)
|
||||
torch.cuda.empty_cache()
|
||||
return results
|
||||
|
||||
|
||||
def is_parallel(model):
|
||||
"""Checks if the model is using Data Parallelism (DP) or Distributed Data Parallelism (DDP)."""
|
||||
return type(model) in (nn.parallel.DataParallel, nn.parallel.DistributedDataParallel)
|
||||
|
||||
|
||||
def de_parallel(model):
|
||||
"""Returns a single-GPU model by removing Data Parallelism (DP) or Distributed Data Parallelism (DDP) if applied."""
|
||||
return model.module if is_parallel(model) else model
|
||||
|
||||
|
||||
def initialize_weights(model):
|
||||
"""Initializes weights of Conv2d, BatchNorm2d, and activations (Hardswish, LeakyReLU, ReLU, ReLU6, SiLU) in the
|
||||
model.
|
||||
"""
|
||||
for m in model.modules():
|
||||
t = type(m)
|
||||
if t is nn.Conv2d:
|
||||
pass # nn.init.kaiming_normal_(m.weight, mode='fan_out', nonlinearity='relu')
|
||||
elif t is nn.BatchNorm2d:
|
||||
m.eps = 1e-3
|
||||
m.momentum = 0.03
|
||||
elif t in [nn.Hardswish, nn.LeakyReLU, nn.ReLU, nn.ReLU6, nn.SiLU]:
|
||||
m.inplace = True
|
||||
|
||||
|
||||
def find_modules(model, mclass=nn.Conv2d):
|
||||
"""Finds and returns list of layer indices in `model.module_list` matching the specified `mclass`."""
|
||||
return [i for i, m in enumerate(model.module_list) if isinstance(m, mclass)]
|
||||
|
||||
|
||||
def sparsity(model):
|
||||
"""Calculates and returns the global sparsity of a model as the ratio of zero-valued parameters to total
|
||||
parameters.
|
||||
"""
|
||||
a, b = 0, 0
|
||||
for p in model.parameters():
|
||||
a += p.numel()
|
||||
b += (p == 0).sum()
|
||||
return b / a
|
||||
|
||||
|
||||
def prune(model, amount=0.3):
|
||||
"""Prunes Conv2d layers in a model to a specified sparsity using L1 unstructured pruning."""
|
||||
import torch.nn.utils.prune as prune
|
||||
|
||||
for name, m in model.named_modules():
|
||||
if isinstance(m, nn.Conv2d):
|
||||
prune.l1_unstructured(m, name="weight", amount=amount) # prune
|
||||
prune.remove(m, "weight") # make permanent
|
||||
LOGGER.info(f"Model pruned to {sparsity(model):.3g} global sparsity")
|
||||
|
||||
|
||||
def fuse_conv_and_bn(conv, bn):
|
||||
"""
|
||||
Fuses Conv2d and BatchNorm2d layers into a single Conv2d layer.
|
||||
|
||||
See https://tehnokv.com/posts/fusing-batchnorm-and-conv/.
|
||||
"""
|
||||
fusedconv = (
|
||||
nn.Conv2d(
|
||||
conv.in_channels,
|
||||
conv.out_channels,
|
||||
kernel_size=conv.kernel_size,
|
||||
stride=conv.stride,
|
||||
padding=conv.padding,
|
||||
dilation=conv.dilation,
|
||||
groups=conv.groups,
|
||||
bias=True,
|
||||
)
|
||||
.requires_grad_(False)
|
||||
.to(conv.weight.device)
|
||||
)
|
||||
|
||||
# Prepare filters
|
||||
w_conv = conv.weight.clone().view(conv.out_channels, -1)
|
||||
w_bn = torch.diag(bn.weight.div(torch.sqrt(bn.eps + bn.running_var)))
|
||||
fusedconv.weight.copy_(torch.mm(w_bn, w_conv).view(fusedconv.weight.shape))
|
||||
|
||||
# Prepare spatial bias
|
||||
b_conv = torch.zeros(conv.weight.size(0), device=conv.weight.device) if conv.bias is None else conv.bias
|
||||
b_bn = bn.bias - bn.weight.mul(bn.running_mean).div(torch.sqrt(bn.running_var + bn.eps))
|
||||
fusedconv.bias.copy_(torch.mm(w_bn, b_conv.reshape(-1, 1)).reshape(-1) + b_bn)
|
||||
|
||||
return fusedconv
|
||||
|
||||
|
||||
def model_info(model, verbose=False, imgsz=640):
|
||||
"""
|
||||
Prints model summary including layers, parameters, gradients, and FLOPs; imgsz may be int or list.
|
||||
|
||||
Example: img_size=640 or img_size=[640, 320]
|
||||
"""
|
||||
n_p = sum(x.numel() for x in model.parameters()) # number parameters
|
||||
n_g = sum(x.numel() for x in model.parameters() if x.requires_grad) # number gradients
|
||||
if verbose:
|
||||
print(f"{'layer':>5} {'name':>40} {'gradient':>9} {'parameters':>12} {'shape':>20} {'mu':>10} {'sigma':>10}")
|
||||
for i, (name, p) in enumerate(model.named_parameters()):
|
||||
name = name.replace("module_list.", "")
|
||||
print(
|
||||
"%5g %40s %9s %12g %20s %10.3g %10.3g"
|
||||
% (i, name, p.requires_grad, p.numel(), list(p.shape), p.mean(), p.std())
|
||||
)
|
||||
|
||||
try: # FLOPs
|
||||
p = next(model.parameters())
|
||||
stride = max(int(model.stride.max()), 32) if hasattr(model, "stride") else 32 # max stride
|
||||
im = torch.empty((1, p.shape[1], stride, stride), device=p.device) # input image in BCHW format
|
||||
flops = thop.profile(deepcopy(model), inputs=(im,), verbose=False)[0] / 1e9 * 2 # stride GFLOPs
|
||||
imgsz = imgsz if isinstance(imgsz, list) else [imgsz, imgsz] # expand if int/float
|
||||
fs = f", {flops * imgsz[0] / stride * imgsz[1] / stride:.1f} GFLOPs" # 640x640 GFLOPs
|
||||
except Exception:
|
||||
fs = ""
|
||||
|
||||
name = Path(model.yaml_file).stem.replace("yolov5", "YOLOv5") if hasattr(model, "yaml_file") else "Model"
|
||||
LOGGER.info(f"{name} summary: {len(list(model.modules()))} layers, {n_p} parameters, {n_g} gradients{fs}")
|
||||
|
||||
|
||||
def scale_img(img, ratio=1.0, same_shape=False, gs=32): # img(16,3,256,416)
|
||||
"""Scales an image tensor `img` of shape (bs,3,y,x) by `ratio`, optionally maintaining the original shape, padded to
|
||||
multiples of `gs`.
|
||||
"""
|
||||
if ratio == 1.0:
|
||||
return img
|
||||
h, w = img.shape[2:]
|
||||
s = (int(h * ratio), int(w * ratio)) # new size
|
||||
img = F.interpolate(img, size=s, mode="bilinear", align_corners=False) # resize
|
||||
if not same_shape: # pad/crop img
|
||||
h, w = (math.ceil(x * ratio / gs) * gs for x in (h, w))
|
||||
return F.pad(img, [0, w - s[1], 0, h - s[0]], value=0.447) # value = imagenet mean
|
||||
|
||||
|
||||
def copy_attr(a, b, include=(), exclude=()):
|
||||
"""Copies attributes from object b to a, optionally filtering with include and exclude lists."""
|
||||
for k, v in b.__dict__.items():
|
||||
if (len(include) and k not in include) or k.startswith("_") or k in exclude:
|
||||
continue
|
||||
else:
|
||||
setattr(a, k, v)
|
||||
|
||||
|
||||
def smart_optimizer(model, name="Adam", lr=0.001, momentum=0.9, decay=1e-5):
|
||||
"""
|
||||
Initializes YOLOv5 smart optimizer with 3 parameter groups for different decay configurations.
|
||||
|
||||
Groups are 0) weights with decay, 1) weights no decay, 2) biases no decay.
|
||||
"""
|
||||
g = [], [], [] # optimizer parameter groups
|
||||
bn = tuple(v for k, v in nn.__dict__.items() if "Norm" in k) # normalization layers, i.e. BatchNorm2d()
|
||||
for v in model.modules():
|
||||
for p_name, p in v.named_parameters(recurse=0):
|
||||
if p_name == "bias": # bias (no decay)
|
||||
g[2].append(p)
|
||||
elif p_name == "weight" and isinstance(v, bn): # weight (no decay)
|
||||
g[1].append(p)
|
||||
else:
|
||||
g[0].append(p) # weight (with decay)
|
||||
|
||||
if name == "Adam":
|
||||
optimizer = torch.optim.Adam(g[2], lr=lr, betas=(momentum, 0.999)) # adjust beta1 to momentum
|
||||
elif name == "AdamW":
|
||||
optimizer = torch.optim.AdamW(g[2], lr=lr, betas=(momentum, 0.999), weight_decay=0.0)
|
||||
elif name == "RMSProp":
|
||||
optimizer = torch.optim.RMSprop(g[2], lr=lr, momentum=momentum)
|
||||
elif name == "SGD":
|
||||
optimizer = torch.optim.SGD(g[2], lr=lr, momentum=momentum, nesterov=True)
|
||||
else:
|
||||
raise NotImplementedError(f"Optimizer {name} not implemented.")
|
||||
|
||||
optimizer.add_param_group({"params": g[0], "weight_decay": decay}) # add g0 with weight_decay
|
||||
optimizer.add_param_group({"params": g[1], "weight_decay": 0.0}) # add g1 (BatchNorm2d weights)
|
||||
LOGGER.info(
|
||||
f"{colorstr('optimizer:')} {type(optimizer).__name__}(lr={lr}) with parameter groups "
|
||||
f"{len(g[1])} weight(decay=0.0), {len(g[0])} weight(decay={decay}), {len(g[2])} bias"
|
||||
)
|
||||
return optimizer
|
||||
|
||||
|
||||
def smart_hub_load(repo="ultralytics/yolov5", model="yolov5s", **kwargs):
|
||||
"""YOLOv5 torch.hub.load() wrapper with smart error handling, adjusting torch arguments for compatibility."""
|
||||
if check_version(torch.__version__, "1.9.1"):
|
||||
kwargs["skip_validation"] = True # validation causes GitHub API rate limit errors
|
||||
if check_version(torch.__version__, "1.12.0"):
|
||||
kwargs["trust_repo"] = True # argument required starting in torch 0.12
|
||||
try:
|
||||
return torch.hub.load(repo, model, **kwargs)
|
||||
except Exception:
|
||||
return torch.hub.load(repo, model, force_reload=True, **kwargs)
|
||||
|
||||
|
||||
def smart_resume(ckpt, optimizer, ema=None, weights="yolov5s.pt", epochs=300, resume=True):
|
||||
"""Resumes training from a checkpoint, updating optimizer, ema, and epochs, with optional resume verification."""
|
||||
best_fitness = 0.0
|
||||
start_epoch = ckpt["epoch"] + 1
|
||||
if ckpt["optimizer"] is not None:
|
||||
optimizer.load_state_dict(ckpt["optimizer"]) # optimizer
|
||||
best_fitness = ckpt["best_fitness"]
|
||||
if ema and ckpt.get("ema"):
|
||||
ema.ema.load_state_dict(ckpt["ema"].float().state_dict()) # EMA
|
||||
ema.updates = ckpt["updates"]
|
||||
if resume:
|
||||
assert start_epoch > 0, (
|
||||
f"{weights} training to {epochs} epochs is finished, nothing to resume.\n"
|
||||
f"Start a new training without --resume, i.e. 'python train.py --weights {weights}'"
|
||||
)
|
||||
LOGGER.info(f"Resuming training from {weights} from epoch {start_epoch} to {epochs} total epochs")
|
||||
if epochs < start_epoch:
|
||||
LOGGER.info(f"{weights} has been trained for {ckpt['epoch']} epochs. Fine-tuning for {epochs} more epochs.")
|
||||
epochs += ckpt["epoch"] # finetune additional epochs
|
||||
return best_fitness, start_epoch, epochs
|
||||
|
||||
|
||||
class EarlyStopping:
|
||||
"""Implements early stopping to halt training when no improvement is observed for a specified number of epochs."""
|
||||
|
||||
def __init__(self, patience=30):
|
||||
"""Initializes simple early stopping mechanism for YOLOv5, with adjustable patience for non-improving epochs."""
|
||||
self.best_fitness = 0.0 # i.e. mAP
|
||||
self.best_epoch = 0
|
||||
self.patience = patience or float("inf") # epochs to wait after fitness stops improving to stop
|
||||
self.possible_stop = False # possible stop may occur next epoch
|
||||
|
||||
def __call__(self, epoch, fitness):
|
||||
"""Evaluates if training should stop based on fitness improvement and patience, returning a boolean."""
|
||||
if fitness >= self.best_fitness: # >= 0 to allow for early zero-fitness stage of training
|
||||
self.best_epoch = epoch
|
||||
self.best_fitness = fitness
|
||||
delta = epoch - self.best_epoch # epochs without improvement
|
||||
self.possible_stop = delta >= (self.patience - 1) # possible stop may occur next epoch
|
||||
stop = delta >= self.patience # stop training if patience exceeded
|
||||
if stop:
|
||||
LOGGER.info(
|
||||
f"Stopping training early as no improvement observed in last {self.patience} epochs. "
|
||||
f"Best results observed at epoch {self.best_epoch}, best model saved as best.pt.\n"
|
||||
f"To update EarlyStopping(patience={self.patience}) pass a new patience value, "
|
||||
f"i.e. `python train.py --patience 300` or use `--patience 0` to disable EarlyStopping."
|
||||
)
|
||||
return stop
|
||||
|
||||
|
||||
class ModelEMA:
|
||||
"""Updated Exponential Moving Average (EMA) from https://github.com/rwightman/pytorch-image-models
|
||||
Keeps a moving average of everything in the model state_dict (parameters and buffers)
|
||||
For EMA details see https://www.tensorflow.org/api_docs/python/tf/train/ExponentialMovingAverage.
|
||||
"""
|
||||
|
||||
def __init__(self, model, decay=0.9999, tau=2000, updates=0):
|
||||
"""Initializes EMA with model parameters, decay rate, tau for decay adjustment, and update count; sets model to
|
||||
evaluation mode.
|
||||
"""
|
||||
self.ema = deepcopy(de_parallel(model)).eval() # FP32 EMA
|
||||
self.updates = updates # number of EMA updates
|
||||
self.decay = lambda x: decay * (1 - math.exp(-x / tau)) # decay exponential ramp (to help early epochs)
|
||||
for p in self.ema.parameters():
|
||||
p.requires_grad_(False)
|
||||
|
||||
def update(self, model):
|
||||
"""Updates the Exponential Moving Average (EMA) parameters based on the current model's parameters."""
|
||||
self.updates += 1
|
||||
d = self.decay(self.updates)
|
||||
|
||||
msd = de_parallel(model).state_dict() # model state_dict
|
||||
for k, v in self.ema.state_dict().items():
|
||||
if v.dtype.is_floating_point: # true for FP16 and FP32
|
||||
v *= d
|
||||
v += (1 - d) * msd[k].detach()
|
||||
# assert v.dtype == msd[k].dtype == torch.float32, f'{k}: EMA {v.dtype} and model {msd[k].dtype} must be FP32'
|
||||
|
||||
def update_attr(self, model, include=(), exclude=("process_group", "reducer")):
|
||||
"""Updates EMA attributes by copying specified attributes from model to EMA, excluding certain attributes by
|
||||
default.
|
||||
"""
|
||||
copy_attr(self.ema, model, include, exclude)
|
||||
@@ -1,90 +0,0 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Utils to interact with the Triton Inference Server."""
|
||||
|
||||
import typing
|
||||
from urllib.parse import urlparse
|
||||
|
||||
import torch
|
||||
|
||||
|
||||
class TritonRemoteModel:
|
||||
"""
|
||||
A wrapper over a model served by the Triton Inference Server.
|
||||
|
||||
It can be configured to communicate over GRPC or HTTP. It accepts Torch Tensors as input and returns them as
|
||||
outputs.
|
||||
"""
|
||||
|
||||
def __init__(self, url: str):
|
||||
"""
|
||||
Keyword Arguments:
|
||||
url: Fully qualified address of the Triton server - for e.g. grpc://localhost:8000.
|
||||
"""
|
||||
parsed_url = urlparse(url)
|
||||
if parsed_url.scheme == "grpc":
|
||||
from tritonclient.grpc import InferenceServerClient, InferInput
|
||||
|
||||
self.client = InferenceServerClient(parsed_url.netloc) # Triton GRPC client
|
||||
model_repository = self.client.get_model_repository_index()
|
||||
self.model_name = model_repository.models[0].name
|
||||
self.metadata = self.client.get_model_metadata(self.model_name, as_json=True)
|
||||
|
||||
def create_input_placeholders() -> typing.List[InferInput]:
|
||||
return [
|
||||
InferInput(i["name"], [int(s) for s in i["shape"]], i["datatype"]) for i in self.metadata["inputs"]
|
||||
]
|
||||
|
||||
else:
|
||||
from tritonclient.http import InferenceServerClient, InferInput
|
||||
|
||||
self.client = InferenceServerClient(parsed_url.netloc) # Triton HTTP client
|
||||
model_repository = self.client.get_model_repository_index()
|
||||
self.model_name = model_repository[0]["name"]
|
||||
self.metadata = self.client.get_model_metadata(self.model_name)
|
||||
|
||||
def create_input_placeholders() -> typing.List[InferInput]:
|
||||
return [
|
||||
InferInput(i["name"], [int(s) for s in i["shape"]], i["datatype"]) for i in self.metadata["inputs"]
|
||||
]
|
||||
|
||||
self._create_input_placeholders_fn = create_input_placeholders
|
||||
|
||||
@property
|
||||
def runtime(self):
|
||||
"""Returns the model runtime."""
|
||||
return self.metadata.get("backend", self.metadata.get("platform"))
|
||||
|
||||
def __call__(self, *args, **kwargs) -> typing.Union[torch.Tensor, typing.Tuple[torch.Tensor, ...]]:
|
||||
"""
|
||||
Invokes the model.
|
||||
|
||||
Parameters can be provided via args or kwargs. args, if provided, are assumed to match the order of inputs of
|
||||
the model. kwargs are matched with the model input names.
|
||||
"""
|
||||
inputs = self._create_inputs(*args, **kwargs)
|
||||
response = self.client.infer(model_name=self.model_name, inputs=inputs)
|
||||
result = []
|
||||
for output in self.metadata["outputs"]:
|
||||
tensor = torch.as_tensor(response.as_numpy(output["name"]))
|
||||
result.append(tensor)
|
||||
return result[0] if len(result) == 1 else result
|
||||
|
||||
def _create_inputs(self, *args, **kwargs):
|
||||
"""Creates input tensors from args or kwargs, not both; raises error if none or both are provided."""
|
||||
args_len, kwargs_len = len(args), len(kwargs)
|
||||
if not args_len and not kwargs_len:
|
||||
raise RuntimeError("No inputs provided.")
|
||||
if args_len and kwargs_len:
|
||||
raise RuntimeError("Cannot specify args and kwargs at the same time")
|
||||
|
||||
placeholders = self._create_input_placeholders_fn()
|
||||
if args_len:
|
||||
if args_len != len(placeholders):
|
||||
raise RuntimeError(f"Expected {len(placeholders)} inputs, got {args_len}.")
|
||||
for input, value in zip(placeholders, args):
|
||||
input.set_data_from_numpy(value.cpu().numpy())
|
||||
else:
|
||||
for input in placeholders:
|
||||
value = kwargs[input.name]
|
||||
input.set_data_from_numpy(value.cpu().numpy())
|
||||
return placeholders
|
||||
Reference in New Issue
Block a user