完成训练模块的转移
This commit is contained in:
476
yolov5/utils/loggers/__init__.py
Normal file
476
yolov5/utils/loggers/__init__.py
Normal file
@@ -0,0 +1,476 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Logging utils."""
|
||||
|
||||
import json
|
||||
import os
|
||||
import warnings
|
||||
from pathlib import Path
|
||||
|
||||
import pkg_resources as pkg
|
||||
import torch
|
||||
|
||||
from utils.general import LOGGER, colorstr, cv2
|
||||
from utils.loggers.clearml.clearml_utils import ClearmlLogger
|
||||
from utils.loggers.wandb.wandb_utils import WandbLogger
|
||||
from utils.plots import plot_images, plot_labels, plot_results
|
||||
from utils.torch_utils import de_parallel
|
||||
|
||||
LOGGERS = ("csv", "tb", "wandb", "clearml", "comet") # *.csv, TensorBoard, Weights & Biases, ClearML
|
||||
RANK = int(os.getenv("RANK", -1))
|
||||
|
||||
try:
|
||||
from torch.utils.tensorboard import SummaryWriter
|
||||
except ImportError:
|
||||
|
||||
def SummaryWriter(*args):
|
||||
"""Fall back to SummaryWriter returning None if TensorBoard is not installed."""
|
||||
return None # None = SummaryWriter(str)
|
||||
|
||||
|
||||
try:
|
||||
import wandb
|
||||
|
||||
assert hasattr(wandb, "__version__") # verify package import not local dir
|
||||
if pkg.parse_version(wandb.__version__) >= pkg.parse_version("0.12.2") and RANK in {0, -1}:
|
||||
try:
|
||||
wandb_login_success = wandb.login(timeout=30)
|
||||
except wandb.errors.UsageError: # known non-TTY terminal issue
|
||||
wandb_login_success = False
|
||||
if not wandb_login_success:
|
||||
wandb = None
|
||||
except (ImportError, AssertionError):
|
||||
wandb = None
|
||||
|
||||
try:
|
||||
import clearml
|
||||
|
||||
assert hasattr(clearml, "__version__") # verify package import not local dir
|
||||
except (ImportError, AssertionError):
|
||||
clearml = None
|
||||
|
||||
try:
|
||||
if RANK in {0, -1}:
|
||||
import comet_ml
|
||||
|
||||
assert hasattr(comet_ml, "__version__") # verify package import not local dir
|
||||
from utils.loggers.comet import CometLogger
|
||||
|
||||
else:
|
||||
comet_ml = None
|
||||
except (ImportError, AssertionError):
|
||||
comet_ml = None
|
||||
|
||||
|
||||
def _json_default(value):
|
||||
"""
|
||||
Format `value` for JSON serialization (e.g. unwrap tensors).
|
||||
|
||||
Fall back to strings.
|
||||
"""
|
||||
if isinstance(value, torch.Tensor):
|
||||
try:
|
||||
value = value.item()
|
||||
except ValueError: # "only one element tensors can be converted to Python scalars"
|
||||
pass
|
||||
return value if isinstance(value, float) else str(value)
|
||||
|
||||
|
||||
class Loggers:
|
||||
"""Initializes and manages various logging utilities for tracking YOLOv5 training and validation metrics."""
|
||||
|
||||
def __init__(self, save_dir=None, weights=None, opt=None, hyp=None, logger=None, include=LOGGERS):
|
||||
"""Initializes loggers for YOLOv5 training and validation metrics, paths, and options."""
|
||||
self.save_dir = save_dir
|
||||
self.weights = weights
|
||||
self.opt = opt
|
||||
self.hyp = hyp
|
||||
self.plots = not opt.noplots # plot results
|
||||
self.logger = logger # for printing results to console
|
||||
self.include = include
|
||||
self.keys = [
|
||||
"train/box_loss",
|
||||
"train/obj_loss",
|
||||
"train/cls_loss", # train loss
|
||||
"metrics/precision",
|
||||
"metrics/recall",
|
||||
"metrics/mAP_0.5",
|
||||
"metrics/mAP_0.5:0.95", # metrics
|
||||
"val/box_loss",
|
||||
"val/obj_loss",
|
||||
"val/cls_loss", # val loss
|
||||
"x/lr0",
|
||||
"x/lr1",
|
||||
"x/lr2",
|
||||
] # params
|
||||
self.best_keys = ["best/epoch", "best/precision", "best/recall", "best/mAP_0.5", "best/mAP_0.5:0.95"]
|
||||
for k in LOGGERS:
|
||||
setattr(self, k, None) # init empty logger dictionary
|
||||
self.csv = True # always log to csv
|
||||
self.ndjson_console = "ndjson_console" in self.include # log ndjson to console
|
||||
self.ndjson_file = "ndjson_file" in self.include # log ndjson to file
|
||||
|
||||
# Messages
|
||||
if not comet_ml:
|
||||
prefix = colorstr("Comet: ")
|
||||
s = f"{prefix}run 'pip install comet_ml' to automatically track and visualize YOLOv5 🚀 runs in Comet"
|
||||
self.logger.info(s)
|
||||
# TensorBoard
|
||||
s = self.save_dir
|
||||
if "tb" in self.include and not self.opt.evolve:
|
||||
prefix = colorstr("TensorBoard: ")
|
||||
self.logger.info(f"{prefix}Start with 'tensorboard --logdir {s.parent}', view at http://localhost:6006/")
|
||||
self.tb = SummaryWriter(str(s))
|
||||
|
||||
# W&B
|
||||
if wandb and "wandb" in self.include:
|
||||
self.opt.hyp = self.hyp # add hyperparameters
|
||||
self.wandb = WandbLogger(self.opt)
|
||||
else:
|
||||
self.wandb = None
|
||||
|
||||
# ClearML
|
||||
if clearml and "clearml" in self.include:
|
||||
try:
|
||||
self.clearml = ClearmlLogger(self.opt, self.hyp)
|
||||
except Exception:
|
||||
self.clearml = None
|
||||
prefix = colorstr("ClearML: ")
|
||||
LOGGER.warning(
|
||||
f"{prefix}WARNING ⚠️ ClearML is installed but not configured, skipping ClearML logging."
|
||||
f" See https://docs.ultralytics.com/yolov5/tutorials/clearml_logging_integration#readme"
|
||||
)
|
||||
|
||||
else:
|
||||
self.clearml = None
|
||||
|
||||
# Comet
|
||||
if comet_ml and "comet" in self.include:
|
||||
if isinstance(self.opt.resume, str) and self.opt.resume.startswith("comet://"):
|
||||
run_id = self.opt.resume.split("/")[-1]
|
||||
self.comet_logger = CometLogger(self.opt, self.hyp, run_id=run_id)
|
||||
|
||||
else:
|
||||
self.comet_logger = CometLogger(self.opt, self.hyp)
|
||||
|
||||
else:
|
||||
self.comet_logger = None
|
||||
|
||||
@property
|
||||
def remote_dataset(self):
|
||||
"""Fetches dataset dictionary from remote logging services like ClearML, Weights & Biases, or Comet ML."""
|
||||
data_dict = None
|
||||
if self.clearml:
|
||||
data_dict = self.clearml.data_dict
|
||||
if self.wandb:
|
||||
data_dict = self.wandb.data_dict
|
||||
if self.comet_logger:
|
||||
data_dict = self.comet_logger.data_dict
|
||||
|
||||
return data_dict
|
||||
|
||||
def on_train_start(self):
|
||||
"""Initializes the training process for Comet ML logger if it's configured."""
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_train_start()
|
||||
|
||||
def on_pretrain_routine_start(self):
|
||||
"""Invokes pre-training routine start hook for Comet ML logger if available."""
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_pretrain_routine_start()
|
||||
|
||||
def on_pretrain_routine_end(self, labels, names):
|
||||
"""Callback that runs at the end of pre-training routine, logging label plots if enabled."""
|
||||
if self.plots:
|
||||
plot_labels(labels, names, self.save_dir)
|
||||
paths = self.save_dir.glob("*labels*.jpg") # training labels
|
||||
if self.wandb:
|
||||
self.wandb.log({"Labels": [wandb.Image(str(x), caption=x.name) for x in paths]})
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_pretrain_routine_end(paths)
|
||||
if self.clearml:
|
||||
for path in paths:
|
||||
self.clearml.log_plot(title=path.stem, plot_path=path)
|
||||
|
||||
def on_train_batch_end(self, model, ni, imgs, targets, paths, vals):
|
||||
"""Logs training batch end events, plots images, and updates external loggers with batch-end data."""
|
||||
log_dict = dict(zip(self.keys[:3], vals))
|
||||
# Callback runs on train batch end
|
||||
# ni: number integrated batches (since train start)
|
||||
if self.plots:
|
||||
if ni < 3:
|
||||
f = self.save_dir / f"train_batch{ni}.jpg" # filename
|
||||
plot_images(imgs, targets, paths, f)
|
||||
if ni == 0 and self.tb and not self.opt.sync_bn:
|
||||
log_tensorboard_graph(self.tb, model, imgsz=(self.opt.imgsz, self.opt.imgsz))
|
||||
if ni == 10 and (self.wandb or self.clearml):
|
||||
files = sorted(self.save_dir.glob("train*.jpg"))
|
||||
if self.wandb:
|
||||
self.wandb.log({"Mosaics": [wandb.Image(str(f), caption=f.name) for f in files if f.exists()]})
|
||||
if self.clearml:
|
||||
self.clearml.log_debug_samples(files, title="Mosaics")
|
||||
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_train_batch_end(log_dict, step=ni)
|
||||
|
||||
def on_train_epoch_end(self, epoch):
|
||||
"""Callback that updates the current epoch in Weights & Biases at the end of a training epoch."""
|
||||
if self.wandb:
|
||||
self.wandb.current_epoch = epoch + 1
|
||||
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_train_epoch_end(epoch)
|
||||
|
||||
def on_val_start(self):
|
||||
"""Callback that signals the start of a validation phase to the Comet logger."""
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_val_start()
|
||||
|
||||
def on_val_image_end(self, pred, predn, path, names, im):
|
||||
"""Callback that logs a validation image and its predictions to WandB or ClearML."""
|
||||
if self.wandb:
|
||||
self.wandb.val_one_image(pred, predn, path, names, im)
|
||||
if self.clearml:
|
||||
self.clearml.log_image_with_boxes(path, pred, names, im)
|
||||
|
||||
def on_val_batch_end(self, batch_i, im, targets, paths, shapes, out):
|
||||
"""Logs validation batch results to Comet ML during training at the end of each validation batch."""
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_val_batch_end(batch_i, im, targets, paths, shapes, out)
|
||||
|
||||
def on_val_end(self, nt, tp, fp, p, r, f1, ap, ap50, ap_class, confusion_matrix):
|
||||
"""Logs validation results to WandB or ClearML at the end of the validation process."""
|
||||
if self.wandb or self.clearml:
|
||||
files = sorted(self.save_dir.glob("val*.jpg"))
|
||||
if self.wandb:
|
||||
self.wandb.log({"Validation": [wandb.Image(str(f), caption=f.name) for f in files]})
|
||||
if self.clearml:
|
||||
self.clearml.log_debug_samples(files, title="Validation")
|
||||
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_val_end(nt, tp, fp, p, r, f1, ap, ap50, ap_class, confusion_matrix)
|
||||
|
||||
def on_fit_epoch_end(self, vals, epoch, best_fitness, fi):
|
||||
"""Callback that logs metrics and saves them to CSV or NDJSON at the end of each fit (train+val) epoch."""
|
||||
x = dict(zip(self.keys, vals))
|
||||
if self.csv:
|
||||
file = self.save_dir / "results.csv"
|
||||
n = len(x) + 1 # number of cols
|
||||
s = "" if file.exists() else (("%20s," * n % tuple(["epoch"] + self.keys)).rstrip(",") + "\n") # add header
|
||||
with open(file, "a") as f:
|
||||
f.write(s + ("%20.5g," * n % tuple([epoch] + vals)).rstrip(",") + "\n")
|
||||
if self.ndjson_console or self.ndjson_file:
|
||||
json_data = json.dumps(dict(epoch=epoch, **x), default=_json_default)
|
||||
if self.ndjson_console:
|
||||
print(json_data)
|
||||
if self.ndjson_file:
|
||||
file = self.save_dir / "results.ndjson"
|
||||
with open(file, "a") as f:
|
||||
print(json_data, file=f)
|
||||
|
||||
if self.tb:
|
||||
for k, v in x.items():
|
||||
self.tb.add_scalar(k, v, epoch)
|
||||
elif self.clearml: # log to ClearML if TensorBoard not used
|
||||
self.clearml.log_scalars(x, epoch)
|
||||
|
||||
if self.wandb:
|
||||
if best_fitness == fi:
|
||||
best_results = [epoch] + vals[3:7]
|
||||
for i, name in enumerate(self.best_keys):
|
||||
self.wandb.wandb_run.summary[name] = best_results[i] # log best results in the summary
|
||||
self.wandb.log(x)
|
||||
self.wandb.end_epoch()
|
||||
|
||||
if self.clearml:
|
||||
self.clearml.current_epoch_logged_images = set() # reset epoch image limit
|
||||
self.clearml.current_epoch += 1
|
||||
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_fit_epoch_end(x, epoch=epoch)
|
||||
|
||||
def on_model_save(self, last, epoch, final_epoch, best_fitness, fi):
|
||||
"""Callback that handles model saving events, logging to Weights & Biases or ClearML if enabled."""
|
||||
if (epoch + 1) % self.opt.save_period == 0 and not final_epoch and self.opt.save_period != -1:
|
||||
if self.wandb:
|
||||
self.wandb.log_model(last.parent, self.opt, epoch, fi, best_model=best_fitness == fi)
|
||||
if self.clearml:
|
||||
self.clearml.task.update_output_model(
|
||||
model_path=str(last), model_name="Latest Model", auto_delete_file=False
|
||||
)
|
||||
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_model_save(last, epoch, final_epoch, best_fitness, fi)
|
||||
|
||||
def on_train_end(self, last, best, epoch, results):
|
||||
"""Callback that runs at the end of training to save plots and log results."""
|
||||
if self.plots:
|
||||
plot_results(file=self.save_dir / "results.csv") # save results.png
|
||||
files = ["results.png", "confusion_matrix.png", *(f"{x}_curve.png" for x in ("F1", "PR", "P", "R"))]
|
||||
files = [(self.save_dir / f) for f in files if (self.save_dir / f).exists()] # filter
|
||||
self.logger.info(f"Results saved to {colorstr('bold', self.save_dir)}")
|
||||
|
||||
if self.tb and not self.clearml: # These images are already captured by ClearML by now, we don't want doubles
|
||||
for f in files:
|
||||
self.tb.add_image(f.stem, cv2.imread(str(f))[..., ::-1], epoch, dataformats="HWC")
|
||||
|
||||
if self.wandb:
|
||||
self.wandb.log(dict(zip(self.keys[3:10], results)))
|
||||
self.wandb.log({"Results": [wandb.Image(str(f), caption=f.name) for f in files]})
|
||||
# Calling wandb.log. TODO: Refactor this into WandbLogger.log_model
|
||||
if not self.opt.evolve:
|
||||
wandb.log_artifact(
|
||||
str(best if best.exists() else last),
|
||||
type="model",
|
||||
name=f"run_{self.wandb.wandb_run.id}_model",
|
||||
aliases=["latest", "best", "stripped"],
|
||||
)
|
||||
self.wandb.finish_run()
|
||||
|
||||
if self.clearml and not self.opt.evolve:
|
||||
self.clearml.log_summary(dict(zip(self.keys[3:10], results)))
|
||||
[self.clearml.log_plot(title=f.stem, plot_path=f) for f in files]
|
||||
self.clearml.log_model(
|
||||
str(best if best.exists() else last), "Best Model" if best.exists() else "Last Model", epoch
|
||||
)
|
||||
|
||||
if self.comet_logger:
|
||||
final_results = dict(zip(self.keys[3:10], results))
|
||||
self.comet_logger.on_train_end(files, self.save_dir, last, best, epoch, final_results)
|
||||
|
||||
def on_params_update(self, params: dict):
|
||||
"""Updates experiment hyperparameters or configurations in WandB, Comet, or ClearML."""
|
||||
if self.wandb:
|
||||
self.wandb.wandb_run.config.update(params, allow_val_change=True)
|
||||
if self.comet_logger:
|
||||
self.comet_logger.on_params_update(params)
|
||||
if self.clearml:
|
||||
self.clearml.task.connect(params)
|
||||
|
||||
|
||||
class GenericLogger:
|
||||
"""
|
||||
YOLOv5 General purpose logger for non-task specific logging
|
||||
Usage: from utils.loggers import GenericLogger; logger = GenericLogger(...).
|
||||
|
||||
Arguments:
|
||||
opt: Run arguments
|
||||
console_logger: Console logger
|
||||
include: loggers to include
|
||||
"""
|
||||
|
||||
def __init__(self, opt, console_logger, include=("tb", "wandb", "clearml")):
|
||||
"""Initializes a generic logger with optional TensorBoard, W&B, and ClearML support."""
|
||||
self.save_dir = Path(opt.save_dir)
|
||||
self.include = include
|
||||
self.console_logger = console_logger
|
||||
self.csv = self.save_dir / "results.csv" # CSV logger
|
||||
if "tb" in self.include:
|
||||
prefix = colorstr("TensorBoard: ")
|
||||
self.console_logger.info(
|
||||
f"{prefix}Start with 'tensorboard --logdir {self.save_dir.parent}', view at http://localhost:6006/"
|
||||
)
|
||||
self.tb = SummaryWriter(str(self.save_dir))
|
||||
|
||||
if wandb and "wandb" in self.include:
|
||||
self.wandb = wandb.init(
|
||||
project=web_project_name(str(opt.project)), name=None if opt.name == "exp" else opt.name, config=opt
|
||||
)
|
||||
else:
|
||||
self.wandb = None
|
||||
|
||||
if clearml and "clearml" in self.include:
|
||||
try:
|
||||
# Hyp is not available in classification mode
|
||||
hyp = {} if "hyp" not in opt else opt.hyp
|
||||
self.clearml = ClearmlLogger(opt, hyp)
|
||||
except Exception:
|
||||
self.clearml = None
|
||||
prefix = colorstr("ClearML: ")
|
||||
LOGGER.warning(
|
||||
f"{prefix}WARNING ⚠️ ClearML is installed but not configured, skipping ClearML logging."
|
||||
f" See https://docs.ultralytics.com/yolov5/tutorials/clearml_logging_integration"
|
||||
)
|
||||
else:
|
||||
self.clearml = None
|
||||
|
||||
def log_metrics(self, metrics, epoch):
|
||||
"""Logs metrics to CSV, TensorBoard, W&B, and ClearML; `metrics` is a dict, `epoch` is an int."""
|
||||
if self.csv:
|
||||
keys, vals = list(metrics.keys()), list(metrics.values())
|
||||
n = len(metrics) + 1 # number of cols
|
||||
s = "" if self.csv.exists() else (("%23s," * n % tuple(["epoch"] + keys)).rstrip(",") + "\n") # header
|
||||
with open(self.csv, "a") as f:
|
||||
f.write(s + ("%23.5g," * n % tuple([epoch] + vals)).rstrip(",") + "\n")
|
||||
|
||||
if self.tb:
|
||||
for k, v in metrics.items():
|
||||
self.tb.add_scalar(k, v, epoch)
|
||||
|
||||
if self.wandb:
|
||||
self.wandb.log(metrics, step=epoch)
|
||||
|
||||
if self.clearml:
|
||||
self.clearml.log_scalars(metrics, epoch)
|
||||
|
||||
def log_images(self, files, name="Images", epoch=0):
|
||||
"""Logs images to all loggers with optional naming and epoch specification."""
|
||||
files = [Path(f) for f in (files if isinstance(files, (tuple, list)) else [files])] # to Path
|
||||
files = [f for f in files if f.exists()] # filter by exists
|
||||
|
||||
if self.tb:
|
||||
for f in files:
|
||||
self.tb.add_image(f.stem, cv2.imread(str(f))[..., ::-1], epoch, dataformats="HWC")
|
||||
|
||||
if self.wandb:
|
||||
self.wandb.log({name: [wandb.Image(str(f), caption=f.name) for f in files]}, step=epoch)
|
||||
|
||||
if self.clearml:
|
||||
if name == "Results":
|
||||
[self.clearml.log_plot(f.stem, f) for f in files]
|
||||
else:
|
||||
self.clearml.log_debug_samples(files, title=name)
|
||||
|
||||
def log_graph(self, model, imgsz=(640, 640)):
|
||||
"""Logs model graph to all configured loggers with specified input image size."""
|
||||
if self.tb:
|
||||
log_tensorboard_graph(self.tb, model, imgsz)
|
||||
|
||||
def log_model(self, model_path, epoch=0, metadata=None):
|
||||
"""Logs the model to all configured loggers with optional epoch and metadata."""
|
||||
if metadata is None:
|
||||
metadata = {}
|
||||
# Log model to all loggers
|
||||
if self.wandb:
|
||||
art = wandb.Artifact(name=f"run_{wandb.run.id}_model", type="model", metadata=metadata)
|
||||
art.add_file(str(model_path))
|
||||
wandb.log_artifact(art)
|
||||
if self.clearml:
|
||||
self.clearml.log_model(model_path=model_path, model_name=model_path.stem)
|
||||
|
||||
def update_params(self, params):
|
||||
"""Updates logged parameters in WandB and/or ClearML if enabled."""
|
||||
if self.wandb:
|
||||
wandb.run.config.update(params, allow_val_change=True)
|
||||
if self.clearml:
|
||||
self.clearml.task.connect(params)
|
||||
|
||||
|
||||
def log_tensorboard_graph(tb, model, imgsz=(640, 640)):
|
||||
"""Logs the model graph to TensorBoard with specified image size and model."""
|
||||
try:
|
||||
p = next(model.parameters()) # for device, type
|
||||
imgsz = (imgsz, imgsz) if isinstance(imgsz, int) else imgsz # expand
|
||||
im = torch.zeros((1, 3, *imgsz)).to(p.device).type_as(p) # input image (WARNING: must be zeros, not empty)
|
||||
with warnings.catch_warnings():
|
||||
warnings.simplefilter("ignore") # suppress jit trace warning
|
||||
tb.add_graph(torch.jit.trace(de_parallel(model), im, strict=False), [])
|
||||
except Exception as e:
|
||||
LOGGER.warning(f"WARNING ⚠️ TensorBoard graph visualization failure {e}")
|
||||
|
||||
|
||||
def web_project_name(project):
|
||||
"""Converts a local project name to a standardized web project name with optional suffixes."""
|
||||
if not project.startswith("runs/train"):
|
||||
return project
|
||||
suffix = "-Classify" if project.endswith("-cls") else "-Segment" if project.endswith("-seg") else ""
|
||||
return f"YOLOv5{suffix}"
|
||||
222
yolov5/utils/loggers/clearml/README.md
Normal file
222
yolov5/utils/loggers/clearml/README.md
Normal file
@@ -0,0 +1,222 @@
|
||||
# ClearML Integration
|
||||
|
||||
<img align="center" src="https://github.com/thepycoder/clearml_screenshots/raw/main/logos_dark.png#gh-light-mode-only" alt="Clear|ML"><img align="center" src="https://github.com/thepycoder/clearml_screenshots/raw/main/logos_light.png#gh-dark-mode-only" alt="Clear|ML">
|
||||
|
||||
## About ClearML
|
||||
|
||||
[ClearML](https://clear.ml/) is an [open-source](https://github.com/clearml/clearml) toolbox designed to save you time ⏱️.
|
||||
|
||||
🔨 Track every YOLOv5 training run in the <b>experiment manager</b>
|
||||
|
||||
🔧 Version and easily access your custom training data with the integrated ClearML <b>Data Versioning Tool</b>
|
||||
|
||||
🔦 <b>Remotely train and monitor</b> your YOLOv5 training runs using ClearML Agent
|
||||
|
||||
🔬 Get the very best mAP using ClearML <b>Hyperparameter Optimization</b>
|
||||
|
||||
🔭 Turn your newly trained <b>YOLOv5 model into an API</b> with just a few commands using ClearML Serving
|
||||
|
||||
And so much more. It's up to you how many of these tools you want to use, you can stick to the experiment manager, or chain them all together into an impressive pipeline!
|
||||
|
||||
|
||||
|
||||
## 🦾 Setting Things Up
|
||||
|
||||
To keep track of your experiments and/or data, ClearML needs to communicate to a server. You have 2 options to get one:
|
||||
|
||||
Either sign up for free to the [ClearML Hosted Service](https://clear.ml/) or you can set up your own server, see [here](https://clear.ml/docs/latest/docs/deploying_clearml/clearml_server). Even the server is open-source, so even if you're dealing with sensitive data, you should be good to go!
|
||||
|
||||
1. Install the `clearml` python package:
|
||||
|
||||
```bash
|
||||
pip install clearml
|
||||
```
|
||||
|
||||
2. Connect the ClearML SDK to the server by [creating credentials](https://app.clear.ml/settings/workspace-configuration) (go right top to Settings -> Workspace -> Create new credentials), then execute the command below and follow the instructions:
|
||||
|
||||
```bash
|
||||
clearml-init
|
||||
```
|
||||
|
||||
That's it! You're done 😎
|
||||
|
||||
## 🚀 Training YOLOv5 With ClearML
|
||||
|
||||
To enable ClearML experiment tracking, simply install the ClearML pip package.
|
||||
|
||||
```bash
|
||||
pip install clearml>=1.2.0
|
||||
```
|
||||
|
||||
This will enable integration with the YOLOv5 training script. Every training run from now on, will be captured and stored by the ClearML experiment manager.
|
||||
|
||||
If you want to change the `project_name` or `task_name`, use the `--project` and `--name` arguments of the `train.py` script, by default the project will be called `YOLOv5` and the task `Training`. PLEASE NOTE: ClearML uses `/` as a delimiter for subprojects, so be careful when using `/` in your project name!
|
||||
|
||||
```bash
|
||||
python train.py --img 640 --batch 16 --epochs 3 --data coco128.yaml --weights yolov5s.pt --cache
|
||||
```
|
||||
|
||||
or with custom project and task name:
|
||||
|
||||
```bash
|
||||
python train.py --project my_project --name my_training --img 640 --batch 16 --epochs 3 --data coco128.yaml --weights yolov5s.pt --cache
|
||||
```
|
||||
|
||||
This will capture:
|
||||
|
||||
- Source code + uncommitted changes
|
||||
- Installed packages
|
||||
- (Hyper)parameters
|
||||
- Model files (use `--save-period n` to save a checkpoint every n epochs)
|
||||
- Console output
|
||||
- Scalars (mAP_0.5, mAP_0.5:0.95, precision, recall, losses, learning rates, ...)
|
||||
- General info such as machine details, runtime, creation date etc.
|
||||
- All produced plots such as label correlogram and confusion matrix
|
||||
- Images with bounding boxes per epoch
|
||||
- Mosaic per epoch
|
||||
- Validation images per epoch
|
||||
- ...
|
||||
|
||||
That's a lot right? 🤯 Now, we can visualize all of this information in the ClearML UI to get an overview of our training progress. Add custom columns to the table view (such as e.g. mAP_0.5) so you can easily sort on the best performing model. Or select multiple experiments and directly compare them!
|
||||
|
||||
There even more we can do with all of this information, like hyperparameter optimization and remote execution, so keep reading if you want to see how that works!
|
||||
|
||||
## 🔗 Dataset Version Management
|
||||
|
||||
Versioning your data separately from your code is generally a good idea and makes it easy to acquire the latest version too. This repository supports supplying a dataset version ID, and it will make sure to get the data if it's not there yet. Next to that, this workflow also saves the used dataset ID as part of the task parameters, so you will always know for sure which data was used in which experiment!
|
||||
|
||||
|
||||
|
||||
### Prepare Your Dataset
|
||||
|
||||
The YOLOv5 repository supports a number of different datasets by using yaml files containing their information. By default datasets are downloaded to the `../datasets` folder in relation to the repository root folder. So if you downloaded the `coco128` dataset using the link in the yaml or with the scripts provided by yolov5, you get this folder structure:
|
||||
|
||||
```
|
||||
..
|
||||
|_ yolov5
|
||||
|_ datasets
|
||||
|_ coco128
|
||||
|_ images
|
||||
|_ labels
|
||||
|_ LICENSE
|
||||
|_ README.txt
|
||||
```
|
||||
|
||||
But this can be any dataset you wish. Feel free to use your own, as long as you keep to this folder structure.
|
||||
|
||||
Next, ⚠️**copy the corresponding yaml file to the root of the dataset folder**⚠️. This yaml files contains the information ClearML will need to properly use the dataset. You can make this yourself too, of course, just follow the structure of the example yamls.
|
||||
|
||||
Basically we need the following keys: `path`, `train`, `test`, `val`, `nc`, `names`.
|
||||
|
||||
```
|
||||
..
|
||||
|_ yolov5
|
||||
|_ datasets
|
||||
|_ coco128
|
||||
|_ images
|
||||
|_ labels
|
||||
|_ coco128.yaml # <---- HERE!
|
||||
|_ LICENSE
|
||||
|_ README.txt
|
||||
```
|
||||
|
||||
### Upload Your Dataset
|
||||
|
||||
To get this dataset into ClearML as a versioned dataset, go to the dataset root folder and run the following command:
|
||||
|
||||
```bash
|
||||
cd coco128
|
||||
clearml-data sync --project YOLOv5 --name coco128 --folder .
|
||||
```
|
||||
|
||||
The command `clearml-data sync` is actually a shorthand command. You could also run these commands one after the other:
|
||||
|
||||
```bash
|
||||
# Optionally add --parent <parent_dataset_id> if you want to base
|
||||
# this version on another dataset version, so no duplicate files are uploaded!
|
||||
clearml-data create --name coco128 --project YOLOv5
|
||||
clearml-data add --files .
|
||||
clearml-data close
|
||||
```
|
||||
|
||||
### Run Training Using A ClearML Dataset
|
||||
|
||||
Now that you have a ClearML dataset, you can very simply use it to train custom YOLOv5 🚀 models!
|
||||
|
||||
```bash
|
||||
python train.py --img 640 --batch 16 --epochs 3 --data clearml://<your_dataset_id> --weights yolov5s.pt --cache
|
||||
```
|
||||
|
||||
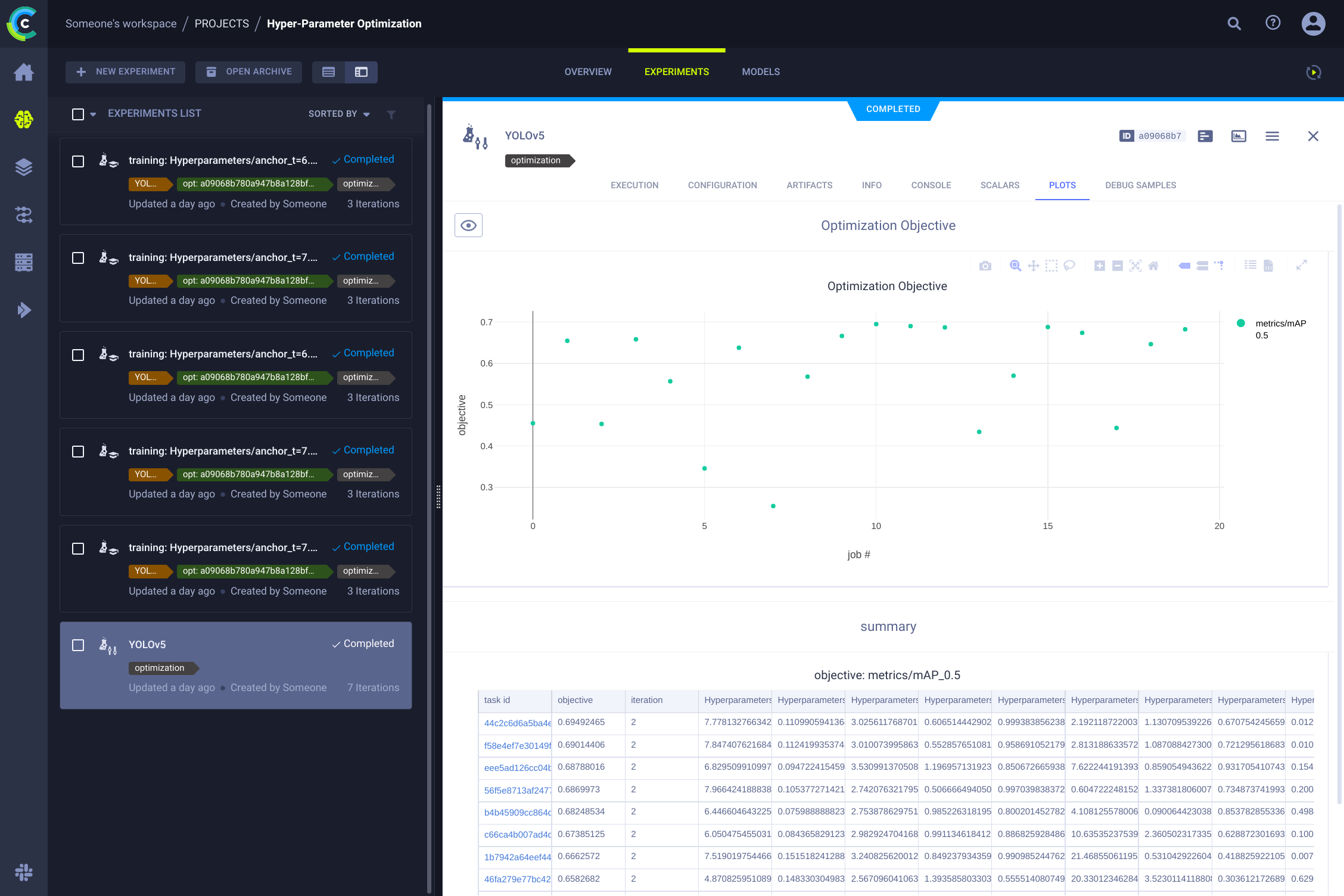
## 👀 Hyperparameter Optimization
|
||||
|
||||
Now that we have our experiments and data versioned, it's time to take a look at what we can build on top!
|
||||
|
||||
Using the code information, installed packages and environment details, the experiment itself is now **completely reproducible**. In fact, ClearML allows you to clone an experiment and even change its parameters. We can then just rerun it with these new parameters automatically, this is basically what HPO does!
|
||||
|
||||
To **run hyperparameter optimization locally**, we've included a pre-made script for you. Just make sure a training task has been run at least once, so it is in the ClearML experiment manager, we will essentially clone it and change its hyperparameters.
|
||||
|
||||
You'll need to fill in the ID of this `template task` in the script found at `utils/loggers/clearml/hpo.py` and then just run it :) You can change `task.execute_locally()` to `task.execute()` to put it in a ClearML queue and have a remote agent work on it instead.
|
||||
|
||||
```bash
|
||||
# To use optuna, install it first, otherwise you can change the optimizer to just be RandomSearch
|
||||
pip install optuna
|
||||
python utils/loggers/clearml/hpo.py
|
||||
```
|
||||
|
||||
|
||||
|
||||
## 🤯 Remote Execution (advanced)
|
||||
|
||||
Running HPO locally is really handy, but what if we want to run our experiments on a remote machine instead? Maybe you have access to a very powerful GPU machine on-site, or you have some budget to use cloud GPUs. This is where the ClearML Agent comes into play. Check out what the agent can do here:
|
||||
|
||||
- [YouTube video](https://www.youtube.com/watch?v=MX3BrXnaULs&feature=youtu.be)
|
||||
- [Documentation](https://clear.ml/docs/latest/docs/clearml_agent)
|
||||
|
||||
In short: every experiment tracked by the experiment manager contains enough information to reproduce it on a different machine (installed packages, uncommitted changes etc.). So a ClearML agent does just that: it listens to a queue for incoming tasks and when it finds one, it recreates the environment and runs it while still reporting scalars, plots etc. to the experiment manager.
|
||||
|
||||
You can turn any machine (a cloud VM, a local GPU machine, your own laptop ... ) into a ClearML agent by simply running:
|
||||
|
||||
```bash
|
||||
clearml-agent daemon --queue <queues_to_listen_to> [--docker]
|
||||
```
|
||||
|
||||
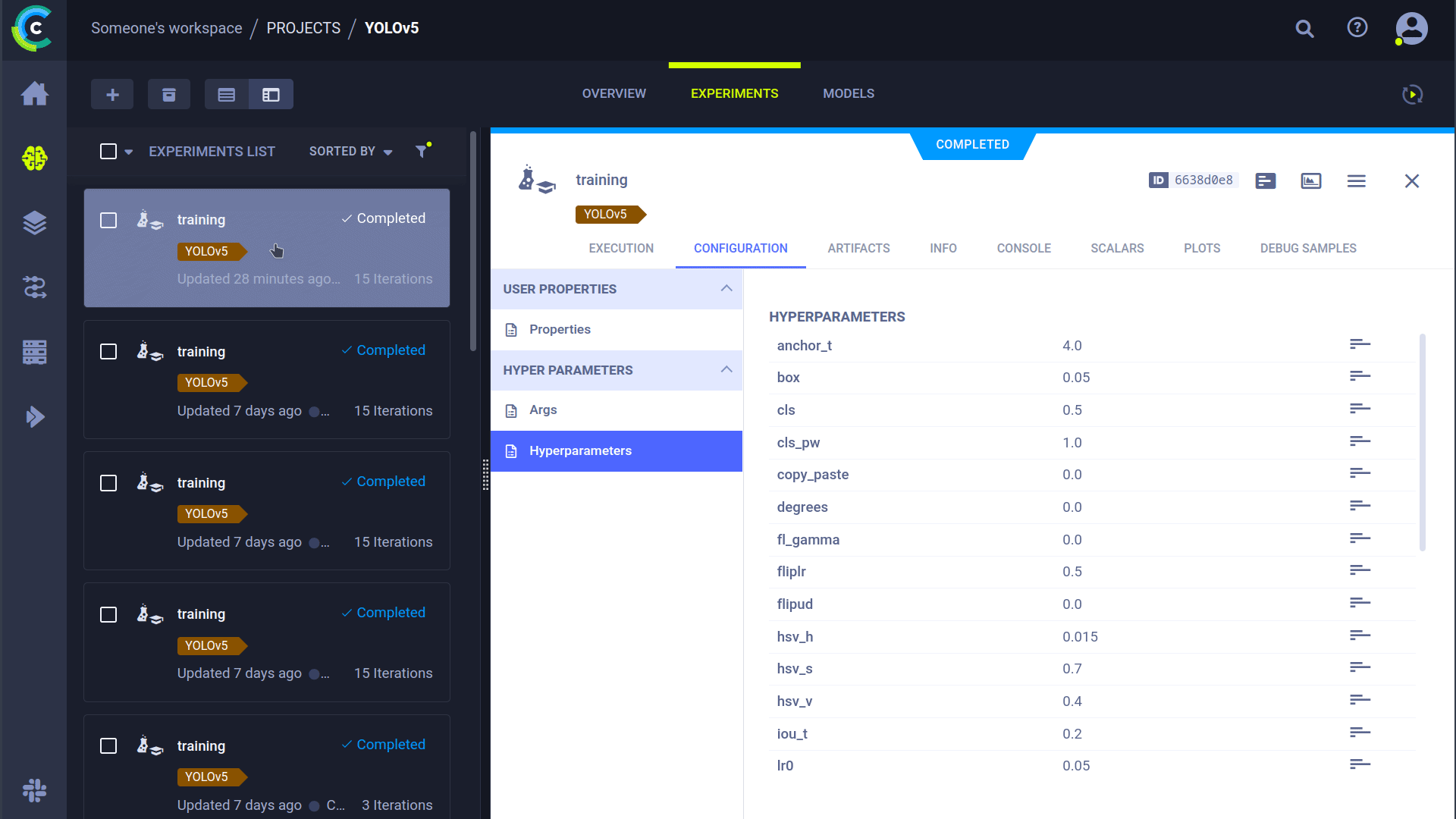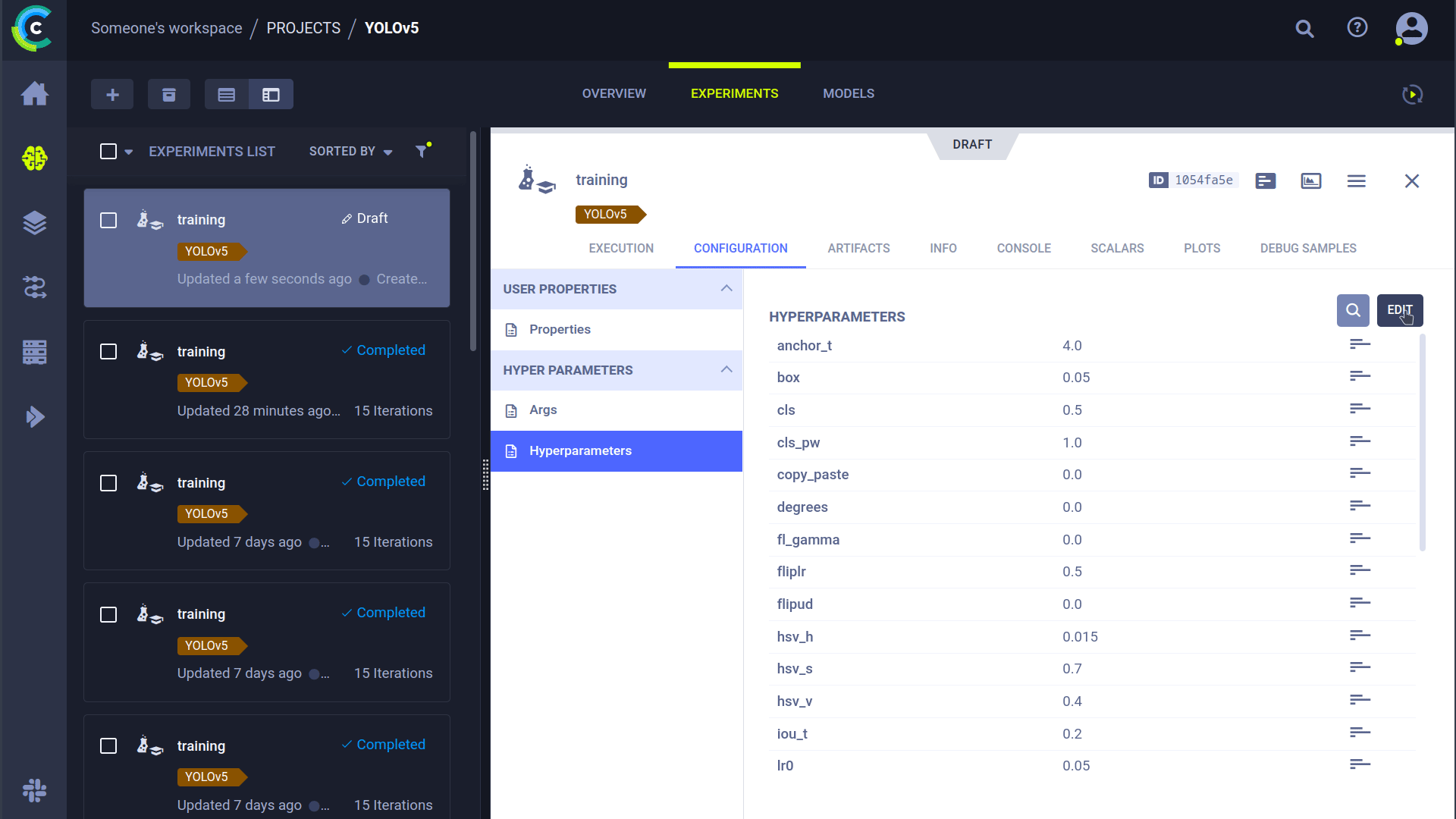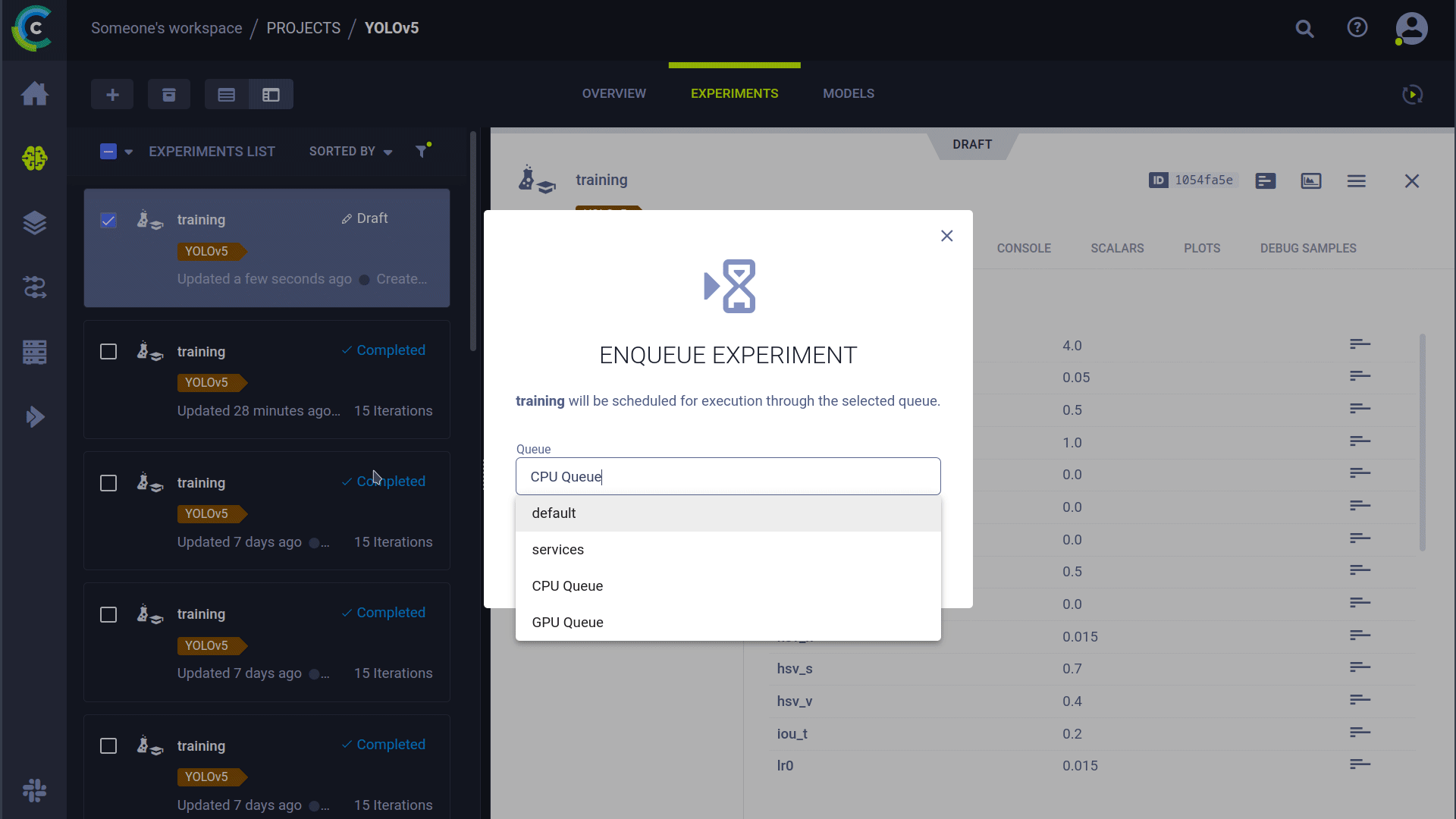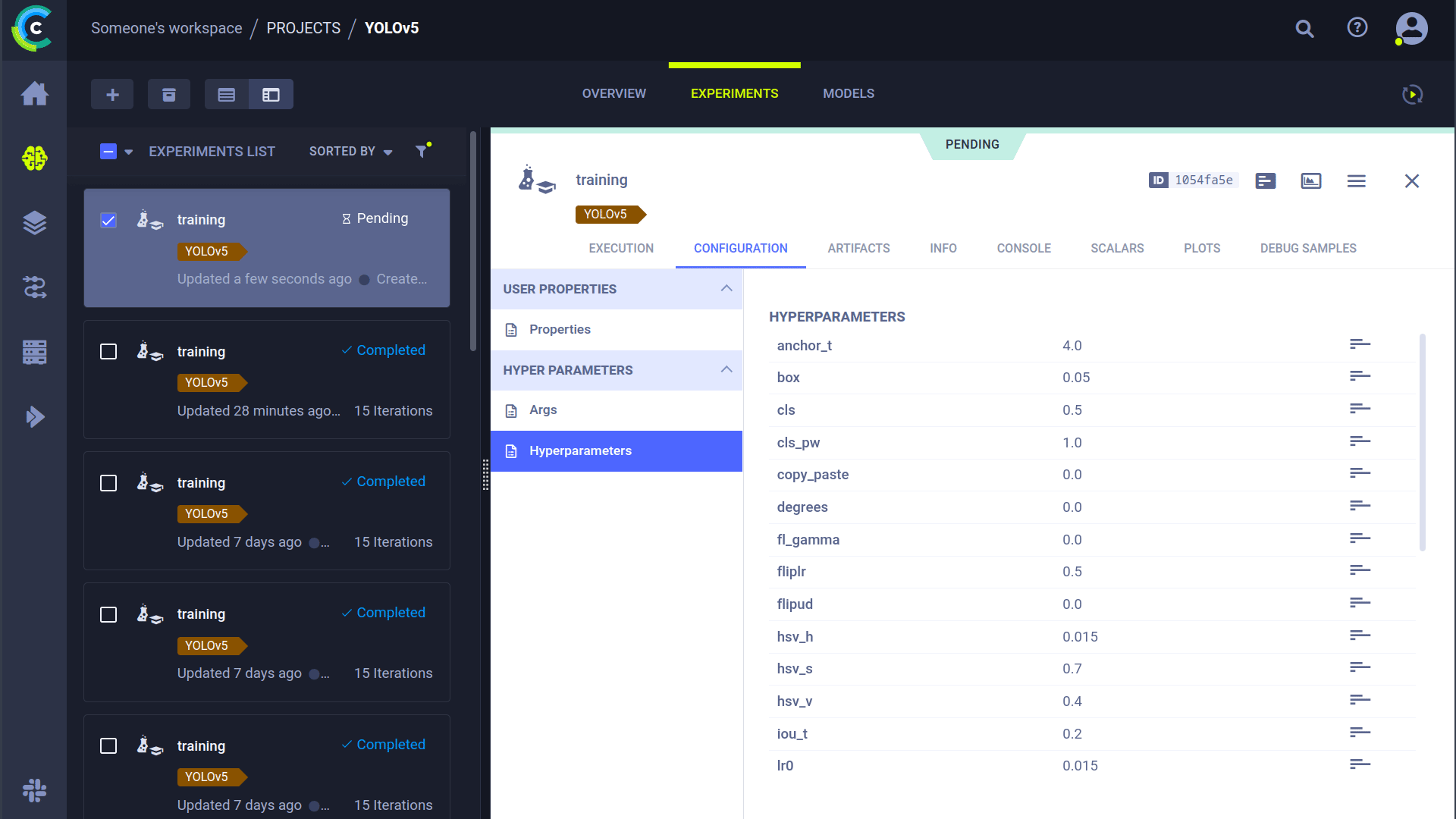
### Cloning, Editing And Enqueuing
|
||||
|
||||
With our agent running, we can give it some work. Remember from the HPO section that we can clone a task and edit the hyperparameters? We can do that from the interface too!
|
||||
|
||||
🪄 Clone the experiment by right-clicking it
|
||||
|
||||
🎯 Edit the hyperparameters to what you wish them to be
|
||||
|
||||
⏳ Enqueue the task to any of the queues by right-clicking it
|
||||
|
||||
|
||||
|
||||
### Executing A Task Remotely
|
||||
|
||||
Now you can clone a task like we explained above, or simply mark your current script by adding `task.execute_remotely()` and on execution it will be put into a queue, for the agent to start working on!
|
||||
|
||||
To run the YOLOv5 training script remotely, all you have to do is add this line to the training.py script after the clearml logger has been instantiated:
|
||||
|
||||
```python
|
||||
# ...
|
||||
# Loggers
|
||||
data_dict = None
|
||||
if RANK in {-1, 0}:
|
||||
loggers = Loggers(save_dir, weights, opt, hyp, LOGGER) # loggers instance
|
||||
if loggers.clearml:
|
||||
loggers.clearml.task.execute_remotely(queue="my_queue") # <------ ADD THIS LINE
|
||||
# Data_dict is either None is user did not choose for ClearML dataset or is filled in by ClearML
|
||||
data_dict = loggers.clearml.data_dict
|
||||
# ...
|
||||
```
|
||||
|
||||
When running the training script after this change, python will run the script up until that line, after which it will package the code and send it to the queue instead!
|
||||
|
||||
### Autoscaling workers
|
||||
|
||||
ClearML comes with autoscalers too! This tool will automatically spin up new remote machines in the cloud of your choice (AWS, GCP, Azure) and turn them into ClearML agents for you whenever there are experiments detected in the queue. Once the tasks are processed, the autoscaler will automatically shut down the remote machines, and you stop paying!
|
||||
|
||||
Check out the autoscalers getting started video below.
|
||||
|
||||
[](https://youtu.be/j4XVMAaUt3E)
|
||||
1
yolov5/utils/loggers/clearml/__init__.py
Normal file
1
yolov5/utils/loggers/clearml/__init__.py
Normal file
@@ -0,0 +1 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
228
yolov5/utils/loggers/clearml/clearml_utils.py
Normal file
228
yolov5/utils/loggers/clearml/clearml_utils.py
Normal file
@@ -0,0 +1,228 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
"""Main Logger class for ClearML experiment tracking."""
|
||||
|
||||
import glob
|
||||
import re
|
||||
from pathlib import Path
|
||||
|
||||
import matplotlib.image as mpimg
|
||||
import matplotlib.pyplot as plt
|
||||
import numpy as np
|
||||
import yaml
|
||||
from ultralytics.utils.plotting import Annotator, colors
|
||||
|
||||
try:
|
||||
import clearml
|
||||
from clearml import Dataset, Task
|
||||
|
||||
assert hasattr(clearml, "__version__") # verify package import not local dir
|
||||
except (ImportError, AssertionError):
|
||||
clearml = None
|
||||
|
||||
|
||||
def construct_dataset(clearml_info_string):
|
||||
"""Load in a clearml dataset and fill the internal data_dict with its contents."""
|
||||
dataset_id = clearml_info_string.replace("clearml://", "")
|
||||
dataset = Dataset.get(dataset_id=dataset_id)
|
||||
dataset_root_path = Path(dataset.get_local_copy())
|
||||
|
||||
# We'll search for the yaml file definition in the dataset
|
||||
yaml_filenames = list(glob.glob(str(dataset_root_path / "*.yaml")) + glob.glob(str(dataset_root_path / "*.yml")))
|
||||
if len(yaml_filenames) > 1:
|
||||
raise ValueError(
|
||||
"More than one yaml file was found in the dataset root, cannot determine which one contains "
|
||||
"the dataset definition this way."
|
||||
)
|
||||
elif not yaml_filenames:
|
||||
raise ValueError(
|
||||
"No yaml definition found in dataset root path, check that there is a correct yaml file "
|
||||
"inside the dataset root path."
|
||||
)
|
||||
with open(yaml_filenames[0]) as f:
|
||||
dataset_definition = yaml.safe_load(f)
|
||||
|
||||
assert set(dataset_definition.keys()).issuperset({"train", "test", "val", "nc", "names"}), (
|
||||
"The right keys were not found in the yaml file, make sure it at least has the following keys: ('train', 'test', 'val', 'nc', 'names')"
|
||||
)
|
||||
|
||||
data_dict = {
|
||||
"train": (
|
||||
str((dataset_root_path / dataset_definition["train"]).resolve()) if dataset_definition["train"] else None
|
||||
)
|
||||
}
|
||||
data_dict["test"] = (
|
||||
str((dataset_root_path / dataset_definition["test"]).resolve()) if dataset_definition["test"] else None
|
||||
)
|
||||
data_dict["val"] = (
|
||||
str((dataset_root_path / dataset_definition["val"]).resolve()) if dataset_definition["val"] else None
|
||||
)
|
||||
data_dict["nc"] = dataset_definition["nc"]
|
||||
data_dict["names"] = dataset_definition["names"]
|
||||
|
||||
return data_dict
|
||||
|
||||
|
||||
class ClearmlLogger:
|
||||
"""
|
||||
Log training runs, datasets, models, and predictions to ClearML.
|
||||
|
||||
This logger sends information to ClearML at app.clear.ml or to your own hosted server. By default, this information
|
||||
includes hyperparameters, system configuration and metrics, model metrics, code information and basic data metrics
|
||||
and analyses.
|
||||
|
||||
By providing additional command line arguments to train.py, datasets, models and predictions can also be logged.
|
||||
"""
|
||||
|
||||
def __init__(self, opt, hyp):
|
||||
"""
|
||||
- Initialize ClearML Task, this object will capture the experiment
|
||||
- Upload dataset version to ClearML Data if opt.upload_dataset is True.
|
||||
|
||||
Arguments:
|
||||
opt (namespace) -- Commandline arguments for this run
|
||||
hyp (dict) -- Hyperparameters for this run
|
||||
|
||||
"""
|
||||
self.current_epoch = 0
|
||||
# Keep tracked of amount of logged images to enforce a limit
|
||||
self.current_epoch_logged_images = set()
|
||||
# Maximum number of images to log to clearML per epoch
|
||||
self.max_imgs_to_log_per_epoch = 16
|
||||
# Get the interval of epochs when bounding box images should be logged
|
||||
# Only for detection task though!
|
||||
if "bbox_interval" in opt:
|
||||
self.bbox_interval = opt.bbox_interval
|
||||
self.clearml = clearml
|
||||
self.task = None
|
||||
self.data_dict = None
|
||||
if self.clearml:
|
||||
self.task = Task.init(
|
||||
project_name="YOLOv5" if str(opt.project).startswith("runs/") else opt.project,
|
||||
task_name=opt.name if opt.name != "exp" else "Training",
|
||||
tags=["YOLOv5"],
|
||||
output_uri=True,
|
||||
reuse_last_task_id=opt.exist_ok,
|
||||
auto_connect_frameworks={"pytorch": False, "matplotlib": False},
|
||||
# We disconnect pytorch auto-detection, because we added manual model save points in the code
|
||||
)
|
||||
# ClearML's hooks will already grab all general parameters
|
||||
# Only the hyperparameters coming from the yaml config file
|
||||
# will have to be added manually!
|
||||
self.task.connect(hyp, name="Hyperparameters")
|
||||
self.task.connect(opt, name="Args")
|
||||
|
||||
# Make sure the code is easily remotely runnable by setting the docker image to use by the remote agent
|
||||
self.task.set_base_docker(
|
||||
"ultralytics/yolov5:latest",
|
||||
docker_arguments='--ipc=host -e="CLEARML_AGENT_SKIP_PYTHON_ENV_INSTALL=1"',
|
||||
docker_setup_bash_script="pip install clearml",
|
||||
)
|
||||
|
||||
# Get ClearML Dataset Version if requested
|
||||
if opt.data.startswith("clearml://"):
|
||||
# data_dict should have the following keys:
|
||||
# names, nc (number of classes), test, train, val (all three relative paths to ../datasets)
|
||||
self.data_dict = construct_dataset(opt.data)
|
||||
# Set data to data_dict because wandb will crash without this information and opt is the best way
|
||||
# to give it to them
|
||||
opt.data = self.data_dict
|
||||
|
||||
def log_scalars(self, metrics, epoch):
|
||||
"""
|
||||
Log scalars/metrics to ClearML.
|
||||
|
||||
Arguments:
|
||||
metrics (dict) Metrics in dict format: {"metrics/mAP": 0.8, ...}
|
||||
epoch (int) iteration number for the current set of metrics
|
||||
"""
|
||||
for k, v in metrics.items():
|
||||
title, series = k.split("/")
|
||||
self.task.get_logger().report_scalar(title, series, v, epoch)
|
||||
|
||||
def log_model(self, model_path, model_name, epoch=0):
|
||||
"""
|
||||
Log model weights to ClearML.
|
||||
|
||||
Arguments:
|
||||
model_path (PosixPath or str) Path to the model weights
|
||||
model_name (str) Name of the model visible in ClearML
|
||||
epoch (int) Iteration / epoch of the model weights
|
||||
"""
|
||||
self.task.update_output_model(
|
||||
model_path=str(model_path), name=model_name, iteration=epoch, auto_delete_file=False
|
||||
)
|
||||
|
||||
def log_summary(self, metrics):
|
||||
"""
|
||||
Log final metrics to a summary table.
|
||||
|
||||
Arguments:
|
||||
metrics (dict) Metrics in dict format: {"metrics/mAP": 0.8, ...}
|
||||
"""
|
||||
for k, v in metrics.items():
|
||||
self.task.get_logger().report_single_value(k, v)
|
||||
|
||||
def log_plot(self, title, plot_path):
|
||||
"""
|
||||
Log image as plot in the plot section of ClearML.
|
||||
|
||||
Arguments:
|
||||
title (str) Title of the plot
|
||||
plot_path (PosixPath or str) Path to the saved image file
|
||||
"""
|
||||
img = mpimg.imread(plot_path)
|
||||
fig = plt.figure()
|
||||
ax = fig.add_axes([0, 0, 1, 1], frameon=False, aspect="auto", xticks=[], yticks=[]) # no ticks
|
||||
ax.imshow(img)
|
||||
|
||||
self.task.get_logger().report_matplotlib_figure(title, "", figure=fig, report_interactive=False)
|
||||
|
||||
def log_debug_samples(self, files, title="Debug Samples"):
|
||||
"""
|
||||
Log files (images) as debug samples in the ClearML task.
|
||||
|
||||
Arguments:
|
||||
files (List(PosixPath)) a list of file paths in PosixPath format
|
||||
title (str) A title that groups together images with the same values
|
||||
"""
|
||||
for f in files:
|
||||
if f.exists():
|
||||
it = re.search(r"_batch(\d+)", f.name)
|
||||
iteration = int(it.groups()[0]) if it else 0
|
||||
self.task.get_logger().report_image(
|
||||
title=title, series=f.name.replace(f"_batch{iteration}", ""), local_path=str(f), iteration=iteration
|
||||
)
|
||||
|
||||
def log_image_with_boxes(self, image_path, boxes, class_names, image, conf_threshold=0.25):
|
||||
"""
|
||||
Draw the bounding boxes on a single image and report the result as a ClearML debug sample.
|
||||
|
||||
Arguments:
|
||||
image_path (PosixPath) the path the original image file
|
||||
boxes (list): list of scaled predictions in the format - [xmin, ymin, xmax, ymax, confidence, class]
|
||||
class_names (dict): dict containing mapping of class int to class name
|
||||
image (Tensor): A torch tensor containing the actual image data
|
||||
"""
|
||||
if (
|
||||
len(self.current_epoch_logged_images) < self.max_imgs_to_log_per_epoch
|
||||
and self.current_epoch >= 0
|
||||
and (self.current_epoch % self.bbox_interval == 0 and image_path not in self.current_epoch_logged_images)
|
||||
):
|
||||
im = np.ascontiguousarray(np.moveaxis(image.mul(255).clamp(0, 255).byte().cpu().numpy(), 0, 2))
|
||||
annotator = Annotator(im=im, pil=True)
|
||||
for i, (conf, class_nr, box) in enumerate(zip(boxes[:, 4], boxes[:, 5], boxes[:, :4])):
|
||||
color = colors(i)
|
||||
|
||||
class_name = class_names[int(class_nr)]
|
||||
confidence_percentage = round(float(conf) * 100, 2)
|
||||
label = f"{class_name}: {confidence_percentage}%"
|
||||
|
||||
if conf > conf_threshold:
|
||||
annotator.rectangle(box.cpu().numpy(), outline=color)
|
||||
annotator.box_label(box.cpu().numpy(), label=label, color=color)
|
||||
|
||||
annotated_image = annotator.result()
|
||||
self.task.get_logger().report_image(
|
||||
title="Bounding Boxes", series=image_path.name, iteration=self.current_epoch, image=annotated_image
|
||||
)
|
||||
self.current_epoch_logged_images.add(image_path)
|
||||
90
yolov5/utils/loggers/clearml/hpo.py
Normal file
90
yolov5/utils/loggers/clearml/hpo.py
Normal file
@@ -0,0 +1,90 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
from clearml import Task
|
||||
|
||||
# Connecting ClearML with the current process,
|
||||
# from here on everything is logged automatically
|
||||
from clearml.automation import HyperParameterOptimizer, UniformParameterRange
|
||||
from clearml.automation.optuna import OptimizerOptuna
|
||||
|
||||
task = Task.init(
|
||||
project_name="Hyper-Parameter Optimization",
|
||||
task_name="YOLOv5",
|
||||
task_type=Task.TaskTypes.optimizer,
|
||||
reuse_last_task_id=False,
|
||||
)
|
||||
|
||||
# Example use case:
|
||||
optimizer = HyperParameterOptimizer(
|
||||
# This is the experiment we want to optimize
|
||||
base_task_id="<your_template_task_id>",
|
||||
# here we define the hyper-parameters to optimize
|
||||
# Notice: The parameter name should exactly match what you see in the UI: <section_name>/<parameter>
|
||||
# For Example, here we see in the base experiment a section Named: "General"
|
||||
# under it a parameter named "batch_size", this becomes "General/batch_size"
|
||||
# If you have `argparse` for example, then arguments will appear under the "Args" section,
|
||||
# and you should instead pass "Args/batch_size"
|
||||
hyper_parameters=[
|
||||
UniformParameterRange("Hyperparameters/lr0", min_value=1e-5, max_value=1e-1),
|
||||
UniformParameterRange("Hyperparameters/lrf", min_value=0.01, max_value=1.0),
|
||||
UniformParameterRange("Hyperparameters/momentum", min_value=0.6, max_value=0.98),
|
||||
UniformParameterRange("Hyperparameters/weight_decay", min_value=0.0, max_value=0.001),
|
||||
UniformParameterRange("Hyperparameters/warmup_epochs", min_value=0.0, max_value=5.0),
|
||||
UniformParameterRange("Hyperparameters/warmup_momentum", min_value=0.0, max_value=0.95),
|
||||
UniformParameterRange("Hyperparameters/warmup_bias_lr", min_value=0.0, max_value=0.2),
|
||||
UniformParameterRange("Hyperparameters/box", min_value=0.02, max_value=0.2),
|
||||
UniformParameterRange("Hyperparameters/cls", min_value=0.2, max_value=4.0),
|
||||
UniformParameterRange("Hyperparameters/cls_pw", min_value=0.5, max_value=2.0),
|
||||
UniformParameterRange("Hyperparameters/obj", min_value=0.2, max_value=4.0),
|
||||
UniformParameterRange("Hyperparameters/obj_pw", min_value=0.5, max_value=2.0),
|
||||
UniformParameterRange("Hyperparameters/iou_t", min_value=0.1, max_value=0.7),
|
||||
UniformParameterRange("Hyperparameters/anchor_t", min_value=2.0, max_value=8.0),
|
||||
UniformParameterRange("Hyperparameters/fl_gamma", min_value=0.0, max_value=4.0),
|
||||
UniformParameterRange("Hyperparameters/hsv_h", min_value=0.0, max_value=0.1),
|
||||
UniformParameterRange("Hyperparameters/hsv_s", min_value=0.0, max_value=0.9),
|
||||
UniformParameterRange("Hyperparameters/hsv_v", min_value=0.0, max_value=0.9),
|
||||
UniformParameterRange("Hyperparameters/degrees", min_value=0.0, max_value=45.0),
|
||||
UniformParameterRange("Hyperparameters/translate", min_value=0.0, max_value=0.9),
|
||||
UniformParameterRange("Hyperparameters/scale", min_value=0.0, max_value=0.9),
|
||||
UniformParameterRange("Hyperparameters/shear", min_value=0.0, max_value=10.0),
|
||||
UniformParameterRange("Hyperparameters/perspective", min_value=0.0, max_value=0.001),
|
||||
UniformParameterRange("Hyperparameters/flipud", min_value=0.0, max_value=1.0),
|
||||
UniformParameterRange("Hyperparameters/fliplr", min_value=0.0, max_value=1.0),
|
||||
UniformParameterRange("Hyperparameters/mosaic", min_value=0.0, max_value=1.0),
|
||||
UniformParameterRange("Hyperparameters/mixup", min_value=0.0, max_value=1.0),
|
||||
UniformParameterRange("Hyperparameters/copy_paste", min_value=0.0, max_value=1.0),
|
||||
],
|
||||
# this is the objective metric we want to maximize/minimize
|
||||
objective_metric_title="metrics",
|
||||
objective_metric_series="mAP_0.5",
|
||||
# now we decide if we want to maximize it or minimize it (accuracy we maximize)
|
||||
objective_metric_sign="max",
|
||||
# let us limit the number of concurrent experiments,
|
||||
# this in turn will make sure we don't bombard the scheduler with experiments.
|
||||
# if we have an auto-scaler connected, this, by proxy, will limit the number of machine
|
||||
max_number_of_concurrent_tasks=1,
|
||||
# this is the optimizer class (actually doing the optimization)
|
||||
# Currently, we can choose from GridSearch, RandomSearch or OptimizerBOHB (Bayesian optimization Hyper-Band)
|
||||
optimizer_class=OptimizerOptuna,
|
||||
# If specified only the top K performing Tasks will be kept, the others will be automatically archived
|
||||
save_top_k_tasks_only=5, # 5,
|
||||
compute_time_limit=None,
|
||||
total_max_jobs=20,
|
||||
min_iteration_per_job=None,
|
||||
max_iteration_per_job=None,
|
||||
)
|
||||
|
||||
# report every 10 seconds, this is way too often, but we are testing here
|
||||
optimizer.set_report_period(10 / 60)
|
||||
# You can also use the line below instead to run all the optimizer tasks locally, without using queues or agent
|
||||
# an_optimizer.start_locally(job_complete_callback=job_complete_callback)
|
||||
# set the time limit for the optimization process (2 hours)
|
||||
optimizer.set_time_limit(in_minutes=120.0)
|
||||
# Start the optimization process in the local environment
|
||||
optimizer.start_locally()
|
||||
# wait until process is done (notice we are controlling the optimization process in the background)
|
||||
optimizer.wait()
|
||||
# make sure background optimization stopped
|
||||
optimizer.stop()
|
||||
|
||||
print("We are done, good bye")
|
||||
250
yolov5/utils/loggers/comet/README.md
Normal file
250
yolov5/utils/loggers/comet/README.md
Normal file
@@ -0,0 +1,250 @@
|
||||
<img src="https://cdn.comet.ml/img/notebook_logo.png">
|
||||
|
||||
# YOLOv5 with Comet
|
||||
|
||||
This guide will cover how to use YOLOv5 with [Comet](https://bit.ly/yolov5-readme-comet2)
|
||||
|
||||
# About Comet
|
||||
|
||||
Comet builds tools that help data scientists, engineers, and team leaders accelerate and optimize machine learning and deep learning models.
|
||||
|
||||
Track and visualize model metrics in real time, save your hyperparameters, datasets, and model checkpoints, and visualize your model predictions with [Comet Custom Panels](https://www.comet.com/docs/v2/guides/comet-dashboard/code-panels/about-panels/?utm_source=yolov5&utm_medium=partner&utm_campaign=partner_yolov5_2022&utm_content=github)! Comet makes sure you never lose track of your work and makes it easy to share results and collaborate across teams of all sizes!
|
||||
|
||||
# Getting Started
|
||||
|
||||
## Install Comet
|
||||
|
||||
```shell
|
||||
pip install comet_ml
|
||||
```
|
||||
|
||||
## Configure Comet Credentials
|
||||
|
||||
There are two ways to configure Comet with YOLOv5.
|
||||
|
||||
You can either set your credentials through environment variables
|
||||
|
||||
**Environment Variables**
|
||||
|
||||
```shell
|
||||
export COMET_API_KEY=<Your Comet API Key>
|
||||
export COMET_PROJECT_NAME=<Your Comet Project Name> # This will default to 'yolov5'
|
||||
```
|
||||
|
||||
Or create a `.comet.config` file in your working directory and set your credentials there.
|
||||
|
||||
**Comet Configuration File**
|
||||
|
||||
```
|
||||
[comet]
|
||||
api_key=<Your Comet API Key>
|
||||
project_name=<Your Comet Project Name> # This will default to 'yolov5'
|
||||
```
|
||||
|
||||
## Run the Training Script
|
||||
|
||||
```shell
|
||||
# Train YOLOv5s on COCO128 for 5 epochs
|
||||
python train.py --img 640 --batch 16 --epochs 5 --data coco128.yaml --weights yolov5s.pt
|
||||
```
|
||||
|
||||
That's it! Comet will automatically log your hyperparameters, command line arguments, training and validation metrics. You can visualize and analyze your runs in the Comet UI
|
||||
|
||||
<img width="1920" alt="yolo-ui" src="https://user-images.githubusercontent.com/26833433/202851203-164e94e1-2238-46dd-91f8-de020e9d6b41.png">
|
||||
|
||||
# Try out an Example!
|
||||
|
||||
Check out an example of a [completed run here](https://www.comet.com/examples/comet-example-yolov5/a0e29e0e9b984e4a822db2a62d0cb357?experiment-tab=chart&showOutliers=true&smoothing=0&transformY=smoothing&xAxis=step&utm_source=yolov5&utm_medium=partner&utm_campaign=partner_yolov5_2022&utm_content=github)
|
||||
|
||||
Or better yet, try it out yourself in this Colab Notebook
|
||||
|
||||
[](https://colab.research.google.com/github/comet-ml/comet-examples/blob/master/integrations/model-training/yolov5/notebooks/Comet_and_YOLOv5.ipynb)
|
||||
|
||||
# Log automatically
|
||||
|
||||
By default, Comet will log the following items
|
||||
|
||||
## Metrics
|
||||
|
||||
- Box Loss, Object Loss, Classification Loss for the training and validation data
|
||||
- mAP_0.5, mAP_0.5:0.95 metrics for the validation data.
|
||||
- Precision and Recall for the validation data
|
||||
|
||||
## Parameters
|
||||
|
||||
- Model Hyperparameters
|
||||
- All parameters passed through the command line options
|
||||
|
||||
## Visualizations
|
||||
|
||||
- Confusion Matrix of the model predictions on the validation data
|
||||
- Plots for the PR and F1 curves across all classes
|
||||
- Correlogram of the Class Labels
|
||||
|
||||
# Configure Comet Logging
|
||||
|
||||
Comet can be configured to log additional data either through command line flags passed to the training script or through environment variables.
|
||||
|
||||
```shell
|
||||
export COMET_MODE=online # Set whether to run Comet in 'online' or 'offline' mode. Defaults to online
|
||||
export COMET_MODEL_NAME=<your model name> #Set the name for the saved model. Defaults to yolov5
|
||||
export COMET_LOG_CONFUSION_MATRIX=false # Set to disable logging a Comet Confusion Matrix. Defaults to true
|
||||
export COMET_MAX_IMAGE_UPLOADS=<number of allowed images to upload to Comet> # Controls how many total image predictions to log to Comet. Defaults to 100.
|
||||
export COMET_LOG_PER_CLASS_METRICS=true # Set to log evaluation metrics for each detected class at the end of training. Defaults to false
|
||||
export COMET_DEFAULT_CHECKPOINT_FILENAME=<your checkpoint filename> # Set this if you would like to resume training from a different checkpoint. Defaults to 'last.pt'
|
||||
export COMET_LOG_BATCH_LEVEL_METRICS=true # Set this if you would like to log training metrics at the batch level. Defaults to false.
|
||||
export COMET_LOG_PREDICTIONS=true # Set this to false to disable logging model predictions
|
||||
```
|
||||
|
||||
## Logging Checkpoints with Comet
|
||||
|
||||
Logging Models to Comet is disabled by default. To enable it, pass the `save-period` argument to the training script. This will save the logged checkpoints to Comet based on the interval value provided by `save-period`
|
||||
|
||||
```shell
|
||||
python train.py \
|
||||
--img 640 \
|
||||
--batch 16 \
|
||||
--epochs 5 \
|
||||
--data coco128.yaml \
|
||||
--weights yolov5s.pt \
|
||||
--save-period 1
|
||||
```
|
||||
|
||||
## Logging Model Predictions
|
||||
|
||||
By default, model predictions (images, ground truth labels and bounding boxes) will be logged to Comet.
|
||||
|
||||
You can control the frequency of logged predictions and the associated images by passing the `bbox_interval` command line argument. Predictions can be visualized using Comet's Object Detection Custom Panel. This frequency corresponds to every Nth batch of data per epoch. In the example below, we are logging every 2nd batch of data for each epoch.
|
||||
|
||||
**Note:** The YOLOv5 validation dataloader will default to a batch size of 32, so you will have to set the logging frequency accordingly.
|
||||
|
||||
Here is an [example project using the Panel](https://www.comet.com/examples/comet-example-yolov5?shareable=YcwMiJaZSXfcEXpGOHDD12vA1&utm_source=yolov5&utm_medium=partner&utm_campaign=partner_yolov5_2022&utm_content=github)
|
||||
|
||||
```shell
|
||||
python train.py \
|
||||
--img 640 \
|
||||
--batch 16 \
|
||||
--epochs 5 \
|
||||
--data coco128.yaml \
|
||||
--weights yolov5s.pt \
|
||||
--bbox_interval 2
|
||||
```
|
||||
|
||||
### Controlling the number of Prediction Images logged to Comet
|
||||
|
||||
When logging predictions from YOLOv5, Comet will log the images associated with each set of predictions. By default a maximum of 100 validation images are logged. You can increase or decrease this number using the `COMET_MAX_IMAGE_UPLOADS` environment variable.
|
||||
|
||||
```shell
|
||||
env COMET_MAX_IMAGE_UPLOADS=200 python train.py \
|
||||
--img 640 \
|
||||
--batch 16 \
|
||||
--epochs 5 \
|
||||
--data coco128.yaml \
|
||||
--weights yolov5s.pt \
|
||||
--bbox_interval 1
|
||||
```
|
||||
|
||||
### Logging Class Level Metrics
|
||||
|
||||
Use the `COMET_LOG_PER_CLASS_METRICS` environment variable to log mAP, precision, recall, f1 for each class.
|
||||
|
||||
```shell
|
||||
env COMET_LOG_PER_CLASS_METRICS=true python train.py \
|
||||
--img 640 \
|
||||
--batch 16 \
|
||||
--epochs 5 \
|
||||
--data coco128.yaml \
|
||||
--weights yolov5s.pt
|
||||
```
|
||||
|
||||
## Uploading a Dataset to Comet Artifacts
|
||||
|
||||
If you would like to store your data using [Comet Artifacts](https://www.comet.com/docs/v2/guides/data-management/using-artifacts/#learn-more?utm_source=yolov5&utm_medium=partner&utm_campaign=partner_yolov5_2022&utm_content=github), you can do so using the `upload_dataset` flag.
|
||||
|
||||
The dataset be organized in the way described in the [YOLOv5 documentation](https://docs.ultralytics.com/yolov5/tutorials/train_custom_data/). The dataset config `yaml` file must follow the same format as that of the `coco128.yaml` file.
|
||||
|
||||
```shell
|
||||
python train.py \
|
||||
--img 640 \
|
||||
--batch 16 \
|
||||
--epochs 5 \
|
||||
--data coco128.yaml \
|
||||
--weights yolov5s.pt \
|
||||
--upload_dataset
|
||||
```
|
||||
|
||||
You can find the uploaded dataset in the Artifacts tab in your Comet Workspace <img width="1073" alt="artifact-1" src="https://user-images.githubusercontent.com/7529846/186929193-162718bf-ec7b-4eb9-8c3b-86b3763ef8ea.png">
|
||||
|
||||
You can preview the data directly in the Comet UI. <img width="1082" alt="artifact-2" src="https://user-images.githubusercontent.com/7529846/186929215-432c36a9-c109-4eb0-944b-84c2786590d6.png">
|
||||
|
||||
Artifacts are versioned and also support adding metadata about the dataset. Comet will automatically log the metadata from your dataset `yaml` file <img width="963" alt="artifact-3" src="https://user-images.githubusercontent.com/7529846/186929256-9d44d6eb-1a19-42de-889a-bcbca3018f2e.png">
|
||||
|
||||
### Using a saved Artifact
|
||||
|
||||
If you would like to use a dataset from Comet Artifacts, set the `path` variable in your dataset `yaml` file to point to the following Artifact resource URL.
|
||||
|
||||
```
|
||||
# contents of artifact.yaml file
|
||||
path: "comet://<workspace name>/<artifact name>:<artifact version or alias>"
|
||||
```
|
||||
|
||||
Then pass this file to your training script in the following way
|
||||
|
||||
```shell
|
||||
python train.py \
|
||||
--img 640 \
|
||||
--batch 16 \
|
||||
--epochs 5 \
|
||||
--data artifact.yaml \
|
||||
--weights yolov5s.pt
|
||||
```
|
||||
|
||||
Artifacts also allow you to track the lineage of data as it flows through your Experimentation workflow. Here you can see a graph that shows you all the experiments that have used your uploaded dataset. <img width="1391" alt="artifact-4" src="https://user-images.githubusercontent.com/7529846/186929264-4c4014fa-fe51-4f3c-a5c5-f6d24649b1b4.png">
|
||||
|
||||
## Resuming a Training Run
|
||||
|
||||
If your training run is interrupted for any reason, e.g. disrupted internet connection, you can resume the run using the `resume` flag and the Comet Run Path.
|
||||
|
||||
The Run Path has the following format `comet://<your workspace name>/<your project name>/<experiment id>`.
|
||||
|
||||
This will restore the run to its state before the interruption, which includes restoring the model from a checkpoint, restoring all hyperparameters and training arguments and downloading Comet dataset Artifacts if they were used in the original run. The resumed run will continue logging to the existing Experiment in the Comet UI
|
||||
|
||||
```shell
|
||||
python train.py \
|
||||
--resume "comet://<your run path>"
|
||||
```
|
||||
|
||||
## Hyperparameter Search with the Comet Optimizer
|
||||
|
||||
YOLOv5 is also integrated with Comet's Optimizer, making is simple to visualize hyperparameter sweeps in the Comet UI.
|
||||
|
||||
### Configuring an Optimizer Sweep
|
||||
|
||||
To configure the Comet Optimizer, you will have to create a JSON file with the information about the sweep. An example file has been provided in `utils/loggers/comet/optimizer_config.json`
|
||||
|
||||
```shell
|
||||
python utils/loggers/comet/hpo.py \
|
||||
--comet_optimizer_config "utils/loggers/comet/optimizer_config.json"
|
||||
```
|
||||
|
||||
The `hpo.py` script accepts the same arguments as `train.py`. If you wish to pass additional arguments to your sweep simply add them after the script.
|
||||
|
||||
```shell
|
||||
python utils/loggers/comet/hpo.py \
|
||||
--comet_optimizer_config "utils/loggers/comet/optimizer_config.json" \
|
||||
--save-period 1 \
|
||||
--bbox_interval 1
|
||||
```
|
||||
|
||||
### Running a Sweep in Parallel
|
||||
|
||||
```shell
|
||||
comet optimizer -j <set number of workers> utils/loggers/comet/hpo.py \
|
||||
utils/loggers/comet/optimizer_config.json"
|
||||
```
|
||||
|
||||
### Visualizing Results
|
||||
|
||||
Comet provides a number of ways to visualize the results of your sweep. Take a look at a [project with a completed sweep here](https://www.comet.com/examples/comet-example-yolov5/view/PrlArHGuuhDTKC1UuBmTtOSXD/panels?utm_source=yolov5&utm_medium=partner&utm_campaign=partner_yolov5_2022&utm_content=github)
|
||||
|
||||
<img width="1626" alt="hyperparameter-yolo" src="https://user-images.githubusercontent.com/7529846/186914869-7dc1de14-583f-4323-967b-c9a66a29e495.png">
|
||||
549
yolov5/utils/loggers/comet/__init__.py
Normal file
549
yolov5/utils/loggers/comet/__init__.py
Normal file
@@ -0,0 +1,549 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
import glob
|
||||
import json
|
||||
import logging
|
||||
import os
|
||||
import sys
|
||||
from pathlib import Path
|
||||
|
||||
logger = logging.getLogger(__name__)
|
||||
|
||||
FILE = Path(__file__).resolve()
|
||||
ROOT = FILE.parents[3] # YOLOv5 root directory
|
||||
if str(ROOT) not in sys.path:
|
||||
sys.path.append(str(ROOT)) # add ROOT to PATH
|
||||
|
||||
try:
|
||||
import comet_ml
|
||||
|
||||
# Project Configuration
|
||||
config = comet_ml.config.get_config()
|
||||
COMET_PROJECT_NAME = config.get_string(os.getenv("COMET_PROJECT_NAME"), "comet.project_name", default="yolov5")
|
||||
except ImportError:
|
||||
comet_ml = None
|
||||
COMET_PROJECT_NAME = None
|
||||
|
||||
import PIL
|
||||
import torch
|
||||
import torchvision.transforms as T
|
||||
import yaml
|
||||
|
||||
from utils.dataloaders import img2label_paths
|
||||
from utils.general import check_dataset, scale_boxes, xywh2xyxy
|
||||
from utils.metrics import box_iou
|
||||
|
||||
COMET_PREFIX = "comet://"
|
||||
|
||||
COMET_MODE = os.getenv("COMET_MODE", "online")
|
||||
|
||||
# Model Saving Settings
|
||||
COMET_MODEL_NAME = os.getenv("COMET_MODEL_NAME", "yolov5")
|
||||
|
||||
# Dataset Artifact Settings
|
||||
COMET_UPLOAD_DATASET = os.getenv("COMET_UPLOAD_DATASET", "false").lower() == "true"
|
||||
|
||||
# Evaluation Settings
|
||||
COMET_LOG_CONFUSION_MATRIX = os.getenv("COMET_LOG_CONFUSION_MATRIX", "true").lower() == "true"
|
||||
COMET_LOG_PREDICTIONS = os.getenv("COMET_LOG_PREDICTIONS", "true").lower() == "true"
|
||||
COMET_MAX_IMAGE_UPLOADS = int(os.getenv("COMET_MAX_IMAGE_UPLOADS", 100))
|
||||
|
||||
# Confusion Matrix Settings
|
||||
CONF_THRES = float(os.getenv("CONF_THRES", 0.001))
|
||||
IOU_THRES = float(os.getenv("IOU_THRES", 0.6))
|
||||
|
||||
# Batch Logging Settings
|
||||
COMET_LOG_BATCH_METRICS = os.getenv("COMET_LOG_BATCH_METRICS", "false").lower() == "true"
|
||||
COMET_BATCH_LOGGING_INTERVAL = os.getenv("COMET_BATCH_LOGGING_INTERVAL", 1)
|
||||
COMET_PREDICTION_LOGGING_INTERVAL = os.getenv("COMET_PREDICTION_LOGGING_INTERVAL", 1)
|
||||
COMET_LOG_PER_CLASS_METRICS = os.getenv("COMET_LOG_PER_CLASS_METRICS", "false").lower() == "true"
|
||||
|
||||
RANK = int(os.getenv("RANK", -1))
|
||||
|
||||
to_pil = T.ToPILImage()
|
||||
|
||||
|
||||
class CometLogger:
|
||||
"""Log metrics, parameters, source code, models and much more with Comet."""
|
||||
|
||||
def __init__(self, opt, hyp, run_id=None, job_type="Training", **experiment_kwargs) -> None:
|
||||
"""Initializes CometLogger with given options, hyperparameters, run ID, job type, and additional experiment
|
||||
arguments.
|
||||
"""
|
||||
self.job_type = job_type
|
||||
self.opt = opt
|
||||
self.hyp = hyp
|
||||
|
||||
# Comet Flags
|
||||
self.comet_mode = COMET_MODE
|
||||
|
||||
self.save_model = opt.save_period > -1
|
||||
self.model_name = COMET_MODEL_NAME
|
||||
|
||||
# Batch Logging Settings
|
||||
self.log_batch_metrics = COMET_LOG_BATCH_METRICS
|
||||
self.comet_log_batch_interval = COMET_BATCH_LOGGING_INTERVAL
|
||||
|
||||
# Dataset Artifact Settings
|
||||
self.upload_dataset = self.opt.upload_dataset or COMET_UPLOAD_DATASET
|
||||
self.resume = self.opt.resume
|
||||
|
||||
self.default_experiment_kwargs = {
|
||||
"log_code": False,
|
||||
"log_env_gpu": True,
|
||||
"log_env_cpu": True,
|
||||
"project_name": COMET_PROJECT_NAME,
|
||||
} | experiment_kwargs
|
||||
self.experiment = self._get_experiment(self.comet_mode, run_id)
|
||||
self.experiment.set_name(self.opt.name)
|
||||
|
||||
self.data_dict = self.check_dataset(self.opt.data)
|
||||
self.class_names = self.data_dict["names"]
|
||||
self.num_classes = self.data_dict["nc"]
|
||||
|
||||
self.logged_images_count = 0
|
||||
self.max_images = COMET_MAX_IMAGE_UPLOADS
|
||||
|
||||
if run_id is None:
|
||||
self.experiment.log_other("Created from", "YOLOv5")
|
||||
if not isinstance(self.experiment, comet_ml.OfflineExperiment):
|
||||
workspace, project_name, experiment_id = self.experiment.url.split("/")[-3:]
|
||||
self.experiment.log_other(
|
||||
"Run Path",
|
||||
f"{workspace}/{project_name}/{experiment_id}",
|
||||
)
|
||||
self.log_parameters(vars(opt))
|
||||
self.log_parameters(self.opt.hyp)
|
||||
self.log_asset_data(
|
||||
self.opt.hyp,
|
||||
name="hyperparameters.json",
|
||||
metadata={"type": "hyp-config-file"},
|
||||
)
|
||||
self.log_asset(
|
||||
f"{self.opt.save_dir}/opt.yaml",
|
||||
metadata={"type": "opt-config-file"},
|
||||
)
|
||||
|
||||
self.comet_log_confusion_matrix = COMET_LOG_CONFUSION_MATRIX
|
||||
|
||||
if hasattr(self.opt, "conf_thres"):
|
||||
self.conf_thres = self.opt.conf_thres
|
||||
else:
|
||||
self.conf_thres = CONF_THRES
|
||||
if hasattr(self.opt, "iou_thres"):
|
||||
self.iou_thres = self.opt.iou_thres
|
||||
else:
|
||||
self.iou_thres = IOU_THRES
|
||||
|
||||
self.log_parameters({"val_iou_threshold": self.iou_thres, "val_conf_threshold": self.conf_thres})
|
||||
|
||||
self.comet_log_predictions = COMET_LOG_PREDICTIONS
|
||||
if self.opt.bbox_interval == -1:
|
||||
self.comet_log_prediction_interval = 1 if self.opt.epochs < 10 else self.opt.epochs // 10
|
||||
else:
|
||||
self.comet_log_prediction_interval = self.opt.bbox_interval
|
||||
|
||||
if self.comet_log_predictions:
|
||||
self.metadata_dict = {}
|
||||
self.logged_image_names = []
|
||||
|
||||
self.comet_log_per_class_metrics = COMET_LOG_PER_CLASS_METRICS
|
||||
|
||||
self.experiment.log_others(
|
||||
{
|
||||
"comet_mode": COMET_MODE,
|
||||
"comet_max_image_uploads": COMET_MAX_IMAGE_UPLOADS,
|
||||
"comet_log_per_class_metrics": COMET_LOG_PER_CLASS_METRICS,
|
||||
"comet_log_batch_metrics": COMET_LOG_BATCH_METRICS,
|
||||
"comet_log_confusion_matrix": COMET_LOG_CONFUSION_MATRIX,
|
||||
"comet_model_name": COMET_MODEL_NAME,
|
||||
}
|
||||
)
|
||||
|
||||
# Check if running the Experiment with the Comet Optimizer
|
||||
if hasattr(self.opt, "comet_optimizer_id"):
|
||||
self.experiment.log_other("optimizer_id", self.opt.comet_optimizer_id)
|
||||
self.experiment.log_other("optimizer_objective", self.opt.comet_optimizer_objective)
|
||||
self.experiment.log_other("optimizer_metric", self.opt.comet_optimizer_metric)
|
||||
self.experiment.log_other("optimizer_parameters", json.dumps(self.hyp))
|
||||
|
||||
def _get_experiment(self, mode, experiment_id=None):
|
||||
"""Returns a new or existing Comet.ml experiment based on mode and optional experiment_id."""
|
||||
if mode == "offline":
|
||||
return (
|
||||
comet_ml.ExistingOfflineExperiment(
|
||||
previous_experiment=experiment_id,
|
||||
**self.default_experiment_kwargs,
|
||||
)
|
||||
if experiment_id is not None
|
||||
else comet_ml.OfflineExperiment(
|
||||
**self.default_experiment_kwargs,
|
||||
)
|
||||
)
|
||||
try:
|
||||
if experiment_id is not None:
|
||||
return comet_ml.ExistingExperiment(
|
||||
previous_experiment=experiment_id,
|
||||
**self.default_experiment_kwargs,
|
||||
)
|
||||
|
||||
return comet_ml.Experiment(**self.default_experiment_kwargs)
|
||||
|
||||
except ValueError:
|
||||
logger.warning(
|
||||
"COMET WARNING: "
|
||||
"Comet credentials have not been set. "
|
||||
"Comet will default to offline logging. "
|
||||
"Please set your credentials to enable online logging."
|
||||
)
|
||||
return self._get_experiment("offline", experiment_id)
|
||||
|
||||
return
|
||||
|
||||
def log_metrics(self, log_dict, **kwargs):
|
||||
"""Logs metrics to the current experiment, accepting a dictionary of metric names and values."""
|
||||
self.experiment.log_metrics(log_dict, **kwargs)
|
||||
|
||||
def log_parameters(self, log_dict, **kwargs):
|
||||
"""Logs parameters to the current experiment, accepting a dictionary of parameter names and values."""
|
||||
self.experiment.log_parameters(log_dict, **kwargs)
|
||||
|
||||
def log_asset(self, asset_path, **kwargs):
|
||||
"""Logs a file or directory as an asset to the current experiment."""
|
||||
self.experiment.log_asset(asset_path, **kwargs)
|
||||
|
||||
def log_asset_data(self, asset, **kwargs):
|
||||
"""Logs in-memory data as an asset to the current experiment, with optional kwargs."""
|
||||
self.experiment.log_asset_data(asset, **kwargs)
|
||||
|
||||
def log_image(self, img, **kwargs):
|
||||
"""Logs an image to the current experiment with optional kwargs."""
|
||||
self.experiment.log_image(img, **kwargs)
|
||||
|
||||
def log_model(self, path, opt, epoch, fitness_score, best_model=False):
|
||||
"""Logs model checkpoint to experiment with path, options, epoch, fitness, and best model flag."""
|
||||
if not self.save_model:
|
||||
return
|
||||
|
||||
model_metadata = {
|
||||
"fitness_score": fitness_score[-1],
|
||||
"epochs_trained": epoch + 1,
|
||||
"save_period": opt.save_period,
|
||||
"total_epochs": opt.epochs,
|
||||
}
|
||||
|
||||
model_files = glob.glob(f"{path}/*.pt")
|
||||
for model_path in model_files:
|
||||
name = Path(model_path).name
|
||||
|
||||
self.experiment.log_model(
|
||||
self.model_name,
|
||||
file_or_folder=model_path,
|
||||
file_name=name,
|
||||
metadata=model_metadata,
|
||||
overwrite=True,
|
||||
)
|
||||
|
||||
def check_dataset(self, data_file):
|
||||
"""Validates the dataset configuration by loading the YAML file specified in `data_file`."""
|
||||
with open(data_file) as f:
|
||||
data_config = yaml.safe_load(f)
|
||||
|
||||
path = data_config.get("path")
|
||||
if path and path.startswith(COMET_PREFIX):
|
||||
path = data_config["path"].replace(COMET_PREFIX, "")
|
||||
return self.download_dataset_artifact(path)
|
||||
self.log_asset(self.opt.data, metadata={"type": "data-config-file"})
|
||||
|
||||
return check_dataset(data_file)
|
||||
|
||||
def log_predictions(self, image, labelsn, path, shape, predn):
|
||||
"""Logs predictions with IOU filtering, given image, labels, path, shape, and predictions."""
|
||||
if self.logged_images_count >= self.max_images:
|
||||
return
|
||||
detections = predn[predn[:, 4] > self.conf_thres]
|
||||
iou = box_iou(labelsn[:, 1:], detections[:, :4])
|
||||
mask, _ = torch.where(iou > self.iou_thres)
|
||||
if len(mask) == 0:
|
||||
return
|
||||
|
||||
filtered_detections = detections[mask]
|
||||
filtered_labels = labelsn[mask]
|
||||
|
||||
image_id = path.split("/")[-1].split(".")[0]
|
||||
image_name = f"{image_id}_curr_epoch_{self.experiment.curr_epoch}"
|
||||
if image_name not in self.logged_image_names:
|
||||
native_scale_image = PIL.Image.open(path)
|
||||
self.log_image(native_scale_image, name=image_name)
|
||||
self.logged_image_names.append(image_name)
|
||||
|
||||
metadata = [
|
||||
{
|
||||
"label": f"{self.class_names[int(cls)]}-gt",
|
||||
"score": 100,
|
||||
"box": {"x": xyxy[0], "y": xyxy[1], "x2": xyxy[2], "y2": xyxy[3]},
|
||||
}
|
||||
for cls, *xyxy in filtered_labels.tolist()
|
||||
]
|
||||
metadata.extend(
|
||||
{
|
||||
"label": f"{self.class_names[int(cls)]}",
|
||||
"score": conf * 100,
|
||||
"box": {"x": xyxy[0], "y": xyxy[1], "x2": xyxy[2], "y2": xyxy[3]},
|
||||
}
|
||||
for *xyxy, conf, cls in filtered_detections.tolist()
|
||||
)
|
||||
self.metadata_dict[image_name] = metadata
|
||||
self.logged_images_count += 1
|
||||
|
||||
return
|
||||
|
||||
def preprocess_prediction(self, image, labels, shape, pred):
|
||||
"""Processes prediction data, resizing labels and adding dataset metadata."""
|
||||
nl, _ = labels.shape[0], pred.shape[0]
|
||||
|
||||
# Predictions
|
||||
if self.opt.single_cls:
|
||||
pred[:, 5] = 0
|
||||
|
||||
predn = pred.clone()
|
||||
scale_boxes(image.shape[1:], predn[:, :4], shape[0], shape[1])
|
||||
|
||||
labelsn = None
|
||||
if nl:
|
||||
tbox = xywh2xyxy(labels[:, 1:5]) # target boxes
|
||||
scale_boxes(image.shape[1:], tbox, shape[0], shape[1]) # native-space labels
|
||||
labelsn = torch.cat((labels[:, 0:1], tbox), 1) # native-space labels
|
||||
scale_boxes(image.shape[1:], predn[:, :4], shape[0], shape[1]) # native-space pred
|
||||
|
||||
return predn, labelsn
|
||||
|
||||
def add_assets_to_artifact(self, artifact, path, asset_path, split):
|
||||
"""Adds image and label assets to a wandb artifact given dataset split and paths."""
|
||||
img_paths = sorted(glob.glob(f"{asset_path}/*"))
|
||||
label_paths = img2label_paths(img_paths)
|
||||
|
||||
for image_file, label_file in zip(img_paths, label_paths):
|
||||
image_logical_path, label_logical_path = map(lambda x: os.path.relpath(x, path), [image_file, label_file])
|
||||
|
||||
try:
|
||||
artifact.add(
|
||||
image_file,
|
||||
logical_path=image_logical_path,
|
||||
metadata={"split": split},
|
||||
)
|
||||
artifact.add(
|
||||
label_file,
|
||||
logical_path=label_logical_path,
|
||||
metadata={"split": split},
|
||||
)
|
||||
except ValueError as e:
|
||||
logger.error("COMET ERROR: Error adding file to Artifact. Skipping file.")
|
||||
logger.error(f"COMET ERROR: {e}")
|
||||
continue
|
||||
|
||||
return artifact
|
||||
|
||||
def upload_dataset_artifact(self):
|
||||
"""Uploads a YOLOv5 dataset as an artifact to the Comet.ml platform."""
|
||||
dataset_name = self.data_dict.get("dataset_name", "yolov5-dataset")
|
||||
path = str((ROOT / Path(self.data_dict["path"])).resolve())
|
||||
|
||||
metadata = self.data_dict.copy()
|
||||
for key in ["train", "val", "test"]:
|
||||
split_path = metadata.get(key)
|
||||
if split_path is not None:
|
||||
metadata[key] = split_path.replace(path, "")
|
||||
|
||||
artifact = comet_ml.Artifact(name=dataset_name, artifact_type="dataset", metadata=metadata)
|
||||
for key in metadata.keys():
|
||||
if key in ["train", "val", "test"]:
|
||||
if isinstance(self.upload_dataset, str) and (key != self.upload_dataset):
|
||||
continue
|
||||
|
||||
asset_path = self.data_dict.get(key)
|
||||
if asset_path is not None:
|
||||
artifact = self.add_assets_to_artifact(artifact, path, asset_path, key)
|
||||
|
||||
self.experiment.log_artifact(artifact)
|
||||
|
||||
return
|
||||
|
||||
def download_dataset_artifact(self, artifact_path):
|
||||
"""Downloads a dataset artifact to a specified directory using the experiment's logged artifact."""
|
||||
logged_artifact = self.experiment.get_artifact(artifact_path)
|
||||
artifact_save_dir = str(Path(self.opt.save_dir) / logged_artifact.name)
|
||||
logged_artifact.download(artifact_save_dir)
|
||||
|
||||
metadata = logged_artifact.metadata
|
||||
data_dict = metadata.copy()
|
||||
data_dict["path"] = artifact_save_dir
|
||||
|
||||
metadata_names = metadata.get("names")
|
||||
if isinstance(metadata_names, dict):
|
||||
data_dict["names"] = {int(k): v for k, v in metadata.get("names").items()}
|
||||
elif isinstance(metadata_names, list):
|
||||
data_dict["names"] = {int(k): v for k, v in zip(range(len(metadata_names)), metadata_names)}
|
||||
else:
|
||||
raise "Invalid 'names' field in dataset yaml file. Please use a list or dictionary"
|
||||
|
||||
return self.update_data_paths(data_dict)
|
||||
|
||||
def update_data_paths(self, data_dict):
|
||||
"""Updates data paths in the dataset dictionary, defaulting 'path' to an empty string if not present."""
|
||||
path = data_dict.get("path", "")
|
||||
|
||||
for split in ["train", "val", "test"]:
|
||||
if data_dict.get(split):
|
||||
split_path = data_dict.get(split)
|
||||
data_dict[split] = (
|
||||
f"{path}/{split_path}" if isinstance(split, str) else [f"{path}/{x}" for x in split_path]
|
||||
)
|
||||
|
||||
return data_dict
|
||||
|
||||
def on_pretrain_routine_end(self, paths):
|
||||
"""Called at the end of pretraining routine to handle paths if training is not being resumed."""
|
||||
if self.opt.resume:
|
||||
return
|
||||
|
||||
for path in paths:
|
||||
self.log_asset(str(path))
|
||||
|
||||
if self.upload_dataset and not self.resume:
|
||||
self.upload_dataset_artifact()
|
||||
|
||||
return
|
||||
|
||||
def on_train_start(self):
|
||||
"""Logs hyperparameters at the start of training."""
|
||||
self.log_parameters(self.hyp)
|
||||
|
||||
def on_train_epoch_start(self):
|
||||
"""Called at the start of each training epoch."""
|
||||
return
|
||||
|
||||
def on_train_epoch_end(self, epoch):
|
||||
"""Updates the current epoch in the experiment tracking at the end of each epoch."""
|
||||
self.experiment.curr_epoch = epoch
|
||||
|
||||
return
|
||||
|
||||
def on_train_batch_start(self):
|
||||
"""Called at the start of each training batch."""
|
||||
return
|
||||
|
||||
def on_train_batch_end(self, log_dict, step):
|
||||
"""Callback function that updates and logs metrics at the end of each training batch if conditions are met."""
|
||||
self.experiment.curr_step = step
|
||||
if self.log_batch_metrics and (step % self.comet_log_batch_interval == 0):
|
||||
self.log_metrics(log_dict, step=step)
|
||||
|
||||
return
|
||||
|
||||
def on_train_end(self, files, save_dir, last, best, epoch, results):
|
||||
"""Logs metadata and optionally saves model files at the end of training."""
|
||||
if self.comet_log_predictions:
|
||||
curr_epoch = self.experiment.curr_epoch
|
||||
self.experiment.log_asset_data(self.metadata_dict, "image-metadata.json", epoch=curr_epoch)
|
||||
|
||||
for f in files:
|
||||
self.log_asset(f, metadata={"epoch": epoch})
|
||||
self.log_asset(f"{save_dir}/results.csv", metadata={"epoch": epoch})
|
||||
|
||||
if not self.opt.evolve:
|
||||
model_path = str(best if best.exists() else last)
|
||||
name = Path(model_path).name
|
||||
if self.save_model:
|
||||
self.experiment.log_model(
|
||||
self.model_name,
|
||||
file_or_folder=model_path,
|
||||
file_name=name,
|
||||
overwrite=True,
|
||||
)
|
||||
|
||||
# Check if running Experiment with Comet Optimizer
|
||||
if hasattr(self.opt, "comet_optimizer_id"):
|
||||
metric = results.get(self.opt.comet_optimizer_metric)
|
||||
self.experiment.log_other("optimizer_metric_value", metric)
|
||||
|
||||
self.finish_run()
|
||||
|
||||
def on_val_start(self):
|
||||
"""Called at the start of validation, currently a placeholder with no functionality."""
|
||||
return
|
||||
|
||||
def on_val_batch_start(self):
|
||||
"""Placeholder called at the start of a validation batch with no current functionality."""
|
||||
return
|
||||
|
||||
def on_val_batch_end(self, batch_i, images, targets, paths, shapes, outputs):
|
||||
"""Callback executed at the end of a validation batch, conditionally logs predictions to Comet ML."""
|
||||
if not (self.comet_log_predictions and ((batch_i + 1) % self.comet_log_prediction_interval == 0)):
|
||||
return
|
||||
|
||||
for si, pred in enumerate(outputs):
|
||||
if len(pred) == 0:
|
||||
continue
|
||||
|
||||
image = images[si]
|
||||
labels = targets[targets[:, 0] == si, 1:]
|
||||
shape = shapes[si]
|
||||
path = paths[si]
|
||||
predn, labelsn = self.preprocess_prediction(image, labels, shape, pred)
|
||||
if labelsn is not None:
|
||||
self.log_predictions(image, labelsn, path, shape, predn)
|
||||
|
||||
return
|
||||
|
||||
def on_val_end(self, nt, tp, fp, p, r, f1, ap, ap50, ap_class, confusion_matrix):
|
||||
"""Logs per-class metrics to Comet.ml after validation if enabled and more than one class exists."""
|
||||
if self.comet_log_per_class_metrics and self.num_classes > 1:
|
||||
for i, c in enumerate(ap_class):
|
||||
class_name = self.class_names[c]
|
||||
self.experiment.log_metrics(
|
||||
{
|
||||
"mAP@.5": ap50[i],
|
||||
"mAP@.5:.95": ap[i],
|
||||
"precision": p[i],
|
||||
"recall": r[i],
|
||||
"f1": f1[i],
|
||||
"true_positives": tp[i],
|
||||
"false_positives": fp[i],
|
||||
"support": nt[c],
|
||||
},
|
||||
prefix=class_name,
|
||||
)
|
||||
|
||||
if self.comet_log_confusion_matrix:
|
||||
epoch = self.experiment.curr_epoch
|
||||
class_names = list(self.class_names.values())
|
||||
class_names.append("background")
|
||||
num_classes = len(class_names)
|
||||
|
||||
self.experiment.log_confusion_matrix(
|
||||
matrix=confusion_matrix.matrix,
|
||||
max_categories=num_classes,
|
||||
labels=class_names,
|
||||
epoch=epoch,
|
||||
column_label="Actual Category",
|
||||
row_label="Predicted Category",
|
||||
file_name=f"confusion-matrix-epoch-{epoch}.json",
|
||||
)
|
||||
|
||||
def on_fit_epoch_end(self, result, epoch):
|
||||
"""Logs metrics at the end of each training epoch."""
|
||||
self.log_metrics(result, epoch=epoch)
|
||||
|
||||
def on_model_save(self, last, epoch, final_epoch, best_fitness, fi):
|
||||
"""Callback to save model checkpoints periodically if conditions are met."""
|
||||
if ((epoch + 1) % self.opt.save_period == 0 and not final_epoch) and self.opt.save_period != -1:
|
||||
self.log_model(last.parent, self.opt, epoch, fi, best_model=best_fitness == fi)
|
||||
|
||||
def on_params_update(self, params):
|
||||
"""Logs updated parameters during training."""
|
||||
self.log_parameters(params)
|
||||
|
||||
def finish_run(self):
|
||||
"""Ends the current experiment and logs its completion."""
|
||||
self.experiment.end()
|
||||
151
yolov5/utils/loggers/comet/comet_utils.py
Normal file
151
yolov5/utils/loggers/comet/comet_utils.py
Normal file
@@ -0,0 +1,151 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
import logging
|
||||
import os
|
||||
from urllib.parse import urlparse
|
||||
|
||||
try:
|
||||
import comet_ml
|
||||
except ImportError:
|
||||
comet_ml = None
|
||||
|
||||
import yaml
|
||||
|
||||
logger = logging.getLogger(__name__)
|
||||
|
||||
COMET_PREFIX = "comet://"
|
||||
COMET_MODEL_NAME = os.getenv("COMET_MODEL_NAME", "yolov5")
|
||||
COMET_DEFAULT_CHECKPOINT_FILENAME = os.getenv("COMET_DEFAULT_CHECKPOINT_FILENAME", "last.pt")
|
||||
|
||||
|
||||
def download_model_checkpoint(opt, experiment):
|
||||
"""Downloads YOLOv5 model checkpoint from Comet ML experiment, updating `opt.weights` with download path."""
|
||||
model_dir = f"{opt.project}/{experiment.name}"
|
||||
os.makedirs(model_dir, exist_ok=True)
|
||||
|
||||
model_name = COMET_MODEL_NAME
|
||||
model_asset_list = experiment.get_model_asset_list(model_name)
|
||||
|
||||
if len(model_asset_list) == 0:
|
||||
logger.error(f"COMET ERROR: No checkpoints found for model name : {model_name}")
|
||||
return
|
||||
|
||||
model_asset_list = sorted(
|
||||
model_asset_list,
|
||||
key=lambda x: x["step"],
|
||||
reverse=True,
|
||||
)
|
||||
logged_checkpoint_map = {asset["fileName"]: asset["assetId"] for asset in model_asset_list}
|
||||
|
||||
resource_url = urlparse(opt.weights)
|
||||
checkpoint_filename = resource_url.query
|
||||
|
||||
if checkpoint_filename:
|
||||
asset_id = logged_checkpoint_map.get(checkpoint_filename)
|
||||
else:
|
||||
asset_id = logged_checkpoint_map.get(COMET_DEFAULT_CHECKPOINT_FILENAME)
|
||||
checkpoint_filename = COMET_DEFAULT_CHECKPOINT_FILENAME
|
||||
|
||||
if asset_id is None:
|
||||
logger.error(f"COMET ERROR: Checkpoint {checkpoint_filename} not found in the given Experiment")
|
||||
return
|
||||
|
||||
try:
|
||||
logger.info(f"COMET INFO: Downloading checkpoint {checkpoint_filename}")
|
||||
asset_filename = checkpoint_filename
|
||||
|
||||
model_binary = experiment.get_asset(asset_id, return_type="binary", stream=False)
|
||||
model_download_path = f"{model_dir}/{asset_filename}"
|
||||
with open(model_download_path, "wb") as f:
|
||||
f.write(model_binary)
|
||||
|
||||
opt.weights = model_download_path
|
||||
|
||||
except Exception as e:
|
||||
logger.warning("COMET WARNING: Unable to download checkpoint from Comet")
|
||||
logger.exception(e)
|
||||
|
||||
|
||||
def set_opt_parameters(opt, experiment):
|
||||
"""
|
||||
Update the opts Namespace with parameters from Comet's ExistingExperiment when resuming a run.
|
||||
|
||||
Args:
|
||||
opt (argparse.Namespace): Namespace of command line options
|
||||
experiment (comet_ml.APIExperiment): Comet API Experiment object
|
||||
"""
|
||||
asset_list = experiment.get_asset_list()
|
||||
resume_string = opt.resume
|
||||
|
||||
for asset in asset_list:
|
||||
if asset["fileName"] == "opt.yaml":
|
||||
asset_id = asset["assetId"]
|
||||
asset_binary = experiment.get_asset(asset_id, return_type="binary", stream=False)
|
||||
opt_dict = yaml.safe_load(asset_binary)
|
||||
for key, value in opt_dict.items():
|
||||
setattr(opt, key, value)
|
||||
opt.resume = resume_string
|
||||
|
||||
# Save hyperparameters to YAML file
|
||||
# Necessary to pass checks in training script
|
||||
save_dir = f"{opt.project}/{experiment.name}"
|
||||
os.makedirs(save_dir, exist_ok=True)
|
||||
|
||||
hyp_yaml_path = f"{save_dir}/hyp.yaml"
|
||||
with open(hyp_yaml_path, "w") as f:
|
||||
yaml.dump(opt.hyp, f)
|
||||
opt.hyp = hyp_yaml_path
|
||||
|
||||
|
||||
def check_comet_weights(opt):
|
||||
"""
|
||||
Downloads model weights from Comet and updates the weights path to point to saved weights location.
|
||||
|
||||
Args:
|
||||
opt (argparse.Namespace): Command Line arguments passed
|
||||
to YOLOv5 training script
|
||||
|
||||
Returns:
|
||||
None/bool: Return True if weights are successfully downloaded
|
||||
else return None
|
||||
"""
|
||||
if comet_ml is None:
|
||||
return
|
||||
|
||||
if isinstance(opt.weights, str) and opt.weights.startswith(COMET_PREFIX):
|
||||
api = comet_ml.API()
|
||||
resource = urlparse(opt.weights)
|
||||
experiment_path = f"{resource.netloc}{resource.path}"
|
||||
experiment = api.get(experiment_path)
|
||||
download_model_checkpoint(opt, experiment)
|
||||
return True
|
||||
|
||||
return None
|
||||
|
||||
|
||||
def check_comet_resume(opt):
|
||||
"""
|
||||
Restores run parameters to its original state based on the model checkpoint and logged Experiment parameters.
|
||||
|
||||
Args:
|
||||
opt (argparse.Namespace): Command Line arguments passed
|
||||
to YOLOv5 training script
|
||||
|
||||
Returns:
|
||||
None/bool: Return True if the run is restored successfully
|
||||
else return None
|
||||
"""
|
||||
if comet_ml is None:
|
||||
return
|
||||
|
||||
if isinstance(opt.resume, str) and opt.resume.startswith(COMET_PREFIX):
|
||||
api = comet_ml.API()
|
||||
resource = urlparse(opt.resume)
|
||||
experiment_path = f"{resource.netloc}{resource.path}"
|
||||
experiment = api.get(experiment_path)
|
||||
set_opt_parameters(opt, experiment)
|
||||
download_model_checkpoint(opt, experiment)
|
||||
|
||||
return True
|
||||
|
||||
return None
|
||||
126
yolov5/utils/loggers/comet/hpo.py
Normal file
126
yolov5/utils/loggers/comet/hpo.py
Normal file
@@ -0,0 +1,126 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
import argparse
|
||||
import json
|
||||
import logging
|
||||
import os
|
||||
import sys
|
||||
from pathlib import Path
|
||||
|
||||
import comet_ml
|
||||
|
||||
logger = logging.getLogger(__name__)
|
||||
|
||||
FILE = Path(__file__).resolve()
|
||||
ROOT = FILE.parents[3] # YOLOv5 root directory
|
||||
if str(ROOT) not in sys.path:
|
||||
sys.path.append(str(ROOT)) # add ROOT to PATH
|
||||
|
||||
from train import train
|
||||
from utils.callbacks import Callbacks
|
||||
from utils.general import increment_path
|
||||
from utils.torch_utils import select_device
|
||||
|
||||
# Project Configuration
|
||||
config = comet_ml.config.get_config()
|
||||
COMET_PROJECT_NAME = config.get_string(os.getenv("COMET_PROJECT_NAME"), "comet.project_name", default="yolov5")
|
||||
|
||||
|
||||
def get_args(known=False):
|
||||
"""Parses command-line arguments for YOLOv5 training, supporting configuration of weights, data paths,
|
||||
hyperparameters, and more.
|
||||
"""
|
||||
parser = argparse.ArgumentParser()
|
||||
parser.add_argument("--weights", type=str, default=ROOT / "yolov5s.pt", help="initial weights path")
|
||||
parser.add_argument("--cfg", type=str, default="", help="model.yaml path")
|
||||
parser.add_argument("--data", type=str, default=ROOT / "data/coco128.yaml", help="dataset.yaml path")
|
||||
parser.add_argument("--hyp", type=str, default=ROOT / "data/hyps/hyp.scratch-low.yaml", help="hyperparameters path")
|
||||
parser.add_argument("--epochs", type=int, default=300, help="total training epochs")
|
||||
parser.add_argument("--batch-size", type=int, default=16, help="total batch size for all GPUs, -1 for autobatch")
|
||||
parser.add_argument("--imgsz", "--img", "--img-size", type=int, default=640, help="train, val image size (pixels)")
|
||||
parser.add_argument("--rect", action="store_true", help="rectangular training")
|
||||
parser.add_argument("--resume", nargs="?", const=True, default=False, help="resume most recent training")
|
||||
parser.add_argument("--nosave", action="store_true", help="only save final checkpoint")
|
||||
parser.add_argument("--noval", action="store_true", help="only validate final epoch")
|
||||
parser.add_argument("--noautoanchor", action="store_true", help="disable AutoAnchor")
|
||||
parser.add_argument("--noplots", action="store_true", help="save no plot files")
|
||||
parser.add_argument("--evolve", type=int, nargs="?", const=300, help="evolve hyperparameters for x generations")
|
||||
parser.add_argument("--bucket", type=str, default="", help="gsutil bucket")
|
||||
parser.add_argument("--cache", type=str, nargs="?", const="ram", help='--cache images in "ram" (default) or "disk"')
|
||||
parser.add_argument("--image-weights", action="store_true", help="use weighted image selection for training")
|
||||
parser.add_argument("--device", default="", help="cuda device, i.e. 0 or 0,1,2,3 or cpu")
|
||||
parser.add_argument("--multi-scale", action="store_true", help="vary img-size +/- 50%%")
|
||||
parser.add_argument("--single-cls", action="store_true", help="train multi-class data as single-class")
|
||||
parser.add_argument("--optimizer", type=str, choices=["SGD", "Adam", "AdamW"], default="SGD", help="optimizer")
|
||||
parser.add_argument("--sync-bn", action="store_true", help="use SyncBatchNorm, only available in DDP mode")
|
||||
parser.add_argument("--workers", type=int, default=8, help="max dataloader workers (per RANK in DDP mode)")
|
||||
parser.add_argument("--project", default=ROOT / "runs/train", help="save to project/name")
|
||||
parser.add_argument("--name", default="exp", help="save to project/name")
|
||||
parser.add_argument("--exist-ok", action="store_true", help="existing project/name ok, do not increment")
|
||||
parser.add_argument("--quad", action="store_true", help="quad dataloader")
|
||||
parser.add_argument("--cos-lr", action="store_true", help="cosine LR scheduler")
|
||||
parser.add_argument("--label-smoothing", type=float, default=0.0, help="Label smoothing epsilon")
|
||||
parser.add_argument("--patience", type=int, default=100, help="EarlyStopping patience (epochs without improvement)")
|
||||
parser.add_argument("--freeze", nargs="+", type=int, default=[0], help="Freeze layers: backbone=10, first3=0 1 2")
|
||||
parser.add_argument("--save-period", type=int, default=-1, help="Save checkpoint every x epochs (disabled if < 1)")
|
||||
parser.add_argument("--seed", type=int, default=0, help="Global training seed")
|
||||
parser.add_argument("--local_rank", type=int, default=-1, help="Automatic DDP Multi-GPU argument, do not modify")
|
||||
|
||||
# Weights & Biases arguments
|
||||
parser.add_argument("--entity", default=None, help="W&B: Entity")
|
||||
parser.add_argument("--upload_dataset", nargs="?", const=True, default=False, help='W&B: Upload data, "val" option')
|
||||
parser.add_argument("--bbox_interval", type=int, default=-1, help="W&B: Set bounding-box image logging interval")
|
||||
parser.add_argument("--artifact_alias", type=str, default="latest", help="W&B: Version of dataset artifact to use")
|
||||
|
||||
# Comet Arguments
|
||||
parser.add_argument("--comet_optimizer_config", type=str, help="Comet: Path to a Comet Optimizer Config File.")
|
||||
parser.add_argument("--comet_optimizer_id", type=str, help="Comet: ID of the Comet Optimizer sweep.")
|
||||
parser.add_argument("--comet_optimizer_objective", type=str, help="Comet: Set to 'minimize' or 'maximize'.")
|
||||
parser.add_argument("--comet_optimizer_metric", type=str, help="Comet: Metric to Optimize.")
|
||||
parser.add_argument(
|
||||
"--comet_optimizer_workers",
|
||||
type=int,
|
||||
default=1,
|
||||
help="Comet: Number of Parallel Workers to use with the Comet Optimizer.",
|
||||
)
|
||||
|
||||
return parser.parse_known_args()[0] if known else parser.parse_args()
|
||||
|
||||
|
||||
def run(parameters, opt):
|
||||
"""Executes YOLOv5 training with given hyperparameters and options, setting up device and training directories."""
|
||||
hyp_dict = {k: v for k, v in parameters.items() if k not in ["epochs", "batch_size"]}
|
||||
|
||||
opt.save_dir = str(increment_path(Path(opt.project) / opt.name, exist_ok=opt.exist_ok or opt.evolve))
|
||||
opt.batch_size = parameters.get("batch_size")
|
||||
opt.epochs = parameters.get("epochs")
|
||||
|
||||
device = select_device(opt.device, batch_size=opt.batch_size)
|
||||
train(hyp_dict, opt, device, callbacks=Callbacks())
|
||||
|
||||
|
||||
if __name__ == "__main__":
|
||||
opt = get_args(known=True)
|
||||
|
||||
opt.weights = str(opt.weights)
|
||||
opt.cfg = str(opt.cfg)
|
||||
opt.data = str(opt.data)
|
||||
opt.project = str(opt.project)
|
||||
|
||||
optimizer_id = os.getenv("COMET_OPTIMIZER_ID")
|
||||
if optimizer_id is None:
|
||||
with open(opt.comet_optimizer_config) as f:
|
||||
optimizer_config = json.load(f)
|
||||
optimizer = comet_ml.Optimizer(optimizer_config)
|
||||
else:
|
||||
optimizer = comet_ml.Optimizer(optimizer_id)
|
||||
|
||||
opt.comet_optimizer_id = optimizer.id
|
||||
status = optimizer.status()
|
||||
|
||||
opt.comet_optimizer_objective = status["spec"]["objective"]
|
||||
opt.comet_optimizer_metric = status["spec"]["metric"]
|
||||
|
||||
logger.info("COMET INFO: Starting Hyperparameter Sweep")
|
||||
for parameter in optimizer.get_parameters():
|
||||
run(parameter["parameters"], opt)
|
||||
1
yolov5/utils/loggers/wandb/__init__.py
Normal file
1
yolov5/utils/loggers/wandb/__init__.py
Normal file
@@ -0,0 +1 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
210
yolov5/utils/loggers/wandb/wandb_utils.py
Normal file
210
yolov5/utils/loggers/wandb/wandb_utils.py
Normal file
@@ -0,0 +1,210 @@
|
||||
# Ultralytics 🚀 AGPL-3.0 License - https://ultralytics.com/license
|
||||
|
||||
# WARNING ⚠️ wandb is deprecated and will be removed in future release.
|
||||
# See supported integrations at https://github.com/ultralytics/yolov5#integrations
|
||||
|
||||
import logging
|
||||
import os
|
||||
import sys
|
||||
from contextlib import contextmanager
|
||||
from pathlib import Path
|
||||
|
||||
from utils.general import LOGGER, colorstr
|
||||
|
||||
FILE = Path(__file__).resolve()
|
||||
ROOT = FILE.parents[3] # YOLOv5 root directory
|
||||
if str(ROOT) not in sys.path:
|
||||
sys.path.append(str(ROOT)) # add ROOT to PATH
|
||||
RANK = int(os.getenv("RANK", -1))
|
||||
DEPRECATION_WARNING = (
|
||||
f"{colorstr('wandb')}: WARNING ⚠️ wandb is deprecated and will be removed in a future release. "
|
||||
f"See supported integrations at https://github.com/ultralytics/yolov5#integrations."
|
||||
)
|
||||
|
||||
try:
|
||||
import wandb
|
||||
|
||||
assert hasattr(wandb, "__version__") # verify package import not local dir
|
||||
LOGGER.warning(DEPRECATION_WARNING)
|
||||
except (ImportError, AssertionError):
|
||||
wandb = None
|
||||
|
||||
|
||||
class WandbLogger:
|
||||
"""
|
||||
Log training runs, datasets, models, and predictions to Weights & Biases.
|
||||
|
||||
This logger sends information to W&B at wandb.ai. By default, this information includes hyperparameters, system
|
||||
configuration and metrics, model metrics, and basic data metrics and analyses.
|
||||
|
||||
By providing additional command line arguments to train.py, datasets, models and predictions can also be logged.
|
||||
|
||||
For more on how this logger is used, see the Weights & Biases documentation:
|
||||
https://docs.wandb.com/guides/integrations/yolov5
|
||||
"""
|
||||
|
||||
def __init__(self, opt, run_id=None, job_type="Training"):
|
||||
"""
|
||||
- Initialize WandbLogger instance
|
||||
- Upload dataset if opt.upload_dataset is True
|
||||
- Setup training processes if job_type is 'Training'.
|
||||
|
||||
Arguments:
|
||||
opt (namespace) -- Commandline arguments for this run
|
||||
run_id (str) -- Run ID of W&B run to be resumed
|
||||
job_type (str) -- To set the job_type for this run
|
||||
|
||||
"""
|
||||
# Pre-training routine --
|
||||
self.job_type = job_type
|
||||
self.wandb, self.wandb_run = wandb, wandb.run if wandb else None
|
||||
self.val_artifact, self.train_artifact = None, None
|
||||
self.train_artifact_path, self.val_artifact_path = None, None
|
||||
self.result_artifact = None
|
||||
self.val_table, self.result_table = None, None
|
||||
self.max_imgs_to_log = 16
|
||||
self.data_dict = None
|
||||
if self.wandb:
|
||||
self.wandb_run = wandb.run or wandb.init(
|
||||
config=opt,
|
||||
resume="allow",
|
||||
project="YOLOv5" if opt.project == "runs/train" else Path(opt.project).stem,
|
||||
entity=opt.entity,
|
||||
name=opt.name if opt.name != "exp" else None,
|
||||
job_type=job_type,
|
||||
id=run_id,
|
||||
allow_val_change=True,
|
||||
)
|
||||
|
||||
if self.wandb_run and self.job_type == "Training":
|
||||
if isinstance(opt.data, dict):
|
||||
# This means another dataset manager has already processed the dataset info (e.g. ClearML)
|
||||
# and they will have stored the already processed dict in opt.data
|
||||
self.data_dict = opt.data
|
||||
self.setup_training(opt)
|
||||
|
||||
def setup_training(self, opt):
|
||||
"""
|
||||
Setup the necessary processes for training YOLO models:
|
||||
- Attempt to download model checkpoint and dataset artifacts if opt.resume stats with WANDB_ARTIFACT_PREFIX
|
||||
- Update data_dict, to contain info of previous run if resumed and the paths of dataset artifact if downloaded
|
||||
- Setup log_dict, initialize bbox_interval.
|
||||
|
||||
Arguments:
|
||||
opt (namespace) -- commandline arguments for this run
|
||||
|
||||
"""
|
||||
self.log_dict, self.current_epoch = {}, 0
|
||||
self.bbox_interval = opt.bbox_interval
|
||||
if isinstance(opt.resume, str):
|
||||
model_dir, _ = self.download_model_artifact(opt)
|
||||
if model_dir:
|
||||
self.weights = Path(model_dir) / "last.pt"
|
||||
config = self.wandb_run.config
|
||||
opt.weights, opt.save_period, opt.batch_size, opt.bbox_interval, opt.epochs, opt.hyp, opt.imgsz = (
|
||||
str(self.weights),
|
||||
config.save_period,
|
||||
config.batch_size,
|
||||
config.bbox_interval,
|
||||
config.epochs,
|
||||
config.hyp,
|
||||
config.imgsz,
|
||||
)
|
||||
|
||||
if opt.bbox_interval == -1:
|
||||
self.bbox_interval = opt.bbox_interval = (opt.epochs // 10) if opt.epochs > 10 else 1
|
||||
if opt.evolve or opt.noplots:
|
||||
self.bbox_interval = opt.bbox_interval = opt.epochs + 1 # disable bbox_interval
|
||||
|
||||
def log_model(self, path, opt, epoch, fitness_score, best_model=False):
|
||||
"""
|
||||
Log the model checkpoint as W&B artifact.
|
||||
|
||||
Arguments:
|
||||
path (Path) -- Path of directory containing the checkpoints
|
||||
opt (namespace) -- Command line arguments for this run
|
||||
epoch (int) -- Current epoch number
|
||||
fitness_score (float) -- fitness score for current epoch
|
||||
best_model (boolean) -- Boolean representing if the current checkpoint is the best yet.
|
||||
"""
|
||||
model_artifact = wandb.Artifact(
|
||||
f"run_{wandb.run.id}_model",
|
||||
type="model",
|
||||
metadata={
|
||||
"original_url": str(path),
|
||||
"epochs_trained": epoch + 1,
|
||||
"save period": opt.save_period,
|
||||
"project": opt.project,
|
||||
"total_epochs": opt.epochs,
|
||||
"fitness_score": fitness_score,
|
||||
},
|
||||
)
|
||||
model_artifact.add_file(str(path / "last.pt"), name="last.pt")
|
||||
wandb.log_artifact(
|
||||
model_artifact,
|
||||
aliases=[
|
||||
"latest",
|
||||
"last",
|
||||
f"epoch {str(self.current_epoch)}",
|
||||
"best" if best_model else "",
|
||||
],
|
||||
)
|
||||
LOGGER.info(f"Saving model artifact on epoch {epoch + 1}")
|
||||
|
||||
def val_one_image(self, pred, predn, path, names, im):
|
||||
"""Evaluates model prediction for a single image, returning metrics and visualizations."""
|
||||
pass
|
||||
|
||||
def log(self, log_dict):
|
||||
"""
|
||||
Save the metrics to the logging dictionary.
|
||||
|
||||
Arguments:
|
||||
log_dict (Dict) -- metrics/media to be logged in current step
|
||||
"""
|
||||
if self.wandb_run:
|
||||
for key, value in log_dict.items():
|
||||
self.log_dict[key] = value
|
||||
|
||||
def end_epoch(self):
|
||||
"""
|
||||
Commit the log_dict, model artifacts and Tables to W&B and flush the log_dict.
|
||||
|
||||
Arguments:
|
||||
best_result (boolean): Boolean representing if the result of this evaluation is best or not
|
||||
"""
|
||||
if self.wandb_run:
|
||||
with all_logging_disabled():
|
||||
try:
|
||||
wandb.log(self.log_dict)
|
||||
except BaseException as e:
|
||||
LOGGER.info(
|
||||
f"An error occurred in wandb logger. The training will proceed without interruption. More info\n{e}"
|
||||
)
|
||||
self.wandb_run.finish()
|
||||
self.wandb_run = None
|
||||
self.log_dict = {}
|
||||
|
||||
def finish_run(self):
|
||||
"""Log metrics if any and finish the current W&B run."""
|
||||
if self.wandb_run:
|
||||
if self.log_dict:
|
||||
with all_logging_disabled():
|
||||
wandb.log(self.log_dict)
|
||||
wandb.run.finish()
|
||||
LOGGER.warning(DEPRECATION_WARNING)
|
||||
|
||||
|
||||
@contextmanager
|
||||
def all_logging_disabled(highest_level=logging.CRITICAL):
|
||||
"""Source - https://gist.github.com/simon-weber/7853144
|
||||
A context manager that will prevent any logging messages triggered during the body from being processed.
|
||||
:param highest_level: the maximum logging level in use.
|
||||
This would only need to be changed if a custom level greater than CRITICAL is defined.
|
||||
"""
|
||||
previous_level = logging.root.manager.disable
|
||||
logging.disable(highest_level)
|
||||
try:
|
||||
yield
|
||||
finally:
|
||||
logging.disable(previous_level)
|
||||
Reference in New Issue
Block a user